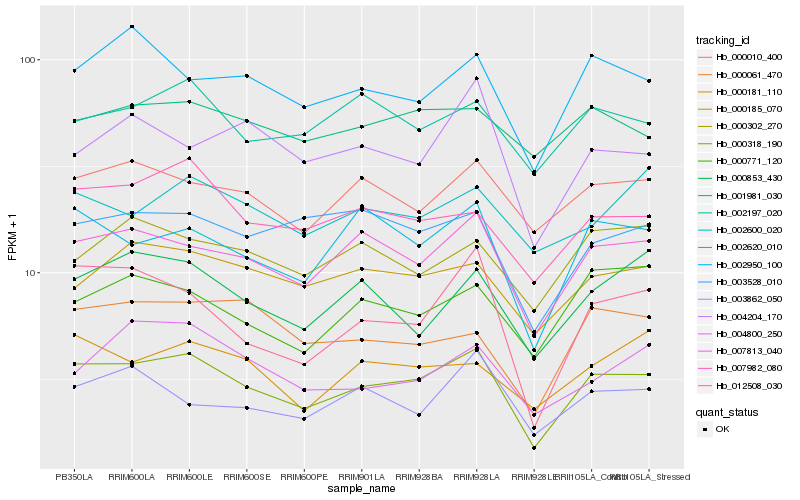
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_190 |
0.0 |
- |
- |
hypothetical protein JCGZ_04889 [Jatropha curcas] |
| 2 |
Hb_007813_040 |
0.0754771617 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 34 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002620_010 |
0.0911977865 |
- |
- |
hypothetical protein POPTR_0004s18130g [Populus trichocarpa] |
| 4 |
Hb_007982_080 |
0.0978446197 |
- |
- |
PREDICTED: uncharacterized protein LOC105636531 [Jatropha curcas] |
| 5 |
Hb_012508_030 |
0.0980371638 |
- |
- |
PREDICTED: uncharacterized protein LOC105636284 [Jatropha curcas] |
| 6 |
Hb_002950_100 |
0.1011270547 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 7 |
Hb_000010_400 |
0.1018841201 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 67 [Jatropha curcas] |
| 8 |
Hb_000185_070 |
0.1044343178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003528_010 |
0.1067362352 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 3R-1 [Jatropha curcas] |
| 10 |
Hb_002197_020 |
0.107880427 |
- |
- |
DNA-directed RNA polymerase family protein [Theobroma cacao] |
| 11 |
Hb_000061_470 |
0.1099895395 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: DNA annealing helicase and endonuclease ZRANB3 [Jatropha curcas] |
| 12 |
Hb_000853_430 |
0.1106242581 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |
| 13 |
Hb_000771_120 |
0.111845135 |
- |
- |
PREDICTED: uncharacterized protein LOC105648352 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000302_270 |
0.1146348782 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_000181_110 |
0.1150795166 |
- |
- |
PREDICTED: uncharacterized protein LOC105637521 [Jatropha curcas] |
| 16 |
Hb_004204_170 |
0.1157008446 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 17 |
Hb_003862_050 |
0.1158795845 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105641042 [Jatropha curcas] |
| 18 |
Hb_001981_030 |
0.1162447106 |
- |
- |
nuclear acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_002600_020 |
0.1168480497 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_004800_250 |
0.1174765574 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |