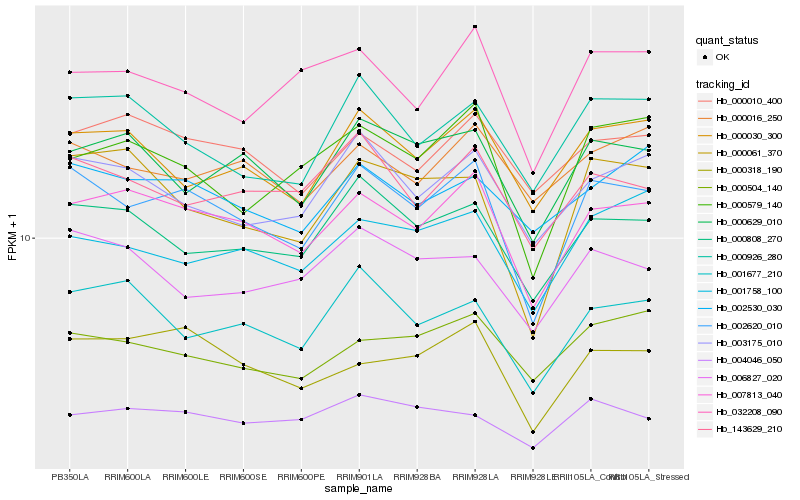
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002620_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s18130g [Populus trichocarpa] |
| 2 |
Hb_007813_040 |
0.0709761017 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 34 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000808_270 |
0.0868062459 |
- |
- |
PREDICTED: lon protease homolog 1, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000318_190 |
0.0911977865 |
- |
- |
hypothetical protein JCGZ_04889 [Jatropha curcas] |
| 5 |
Hb_003175_010 |
0.0942116442 |
- |
- |
PREDICTED: 39S ribosomal protein L4, mitochondrial [Jatropha curcas] |
| 6 |
Hb_143629_210 |
0.0944560381 |
- |
- |
PREDICTED: transcription factor PIF3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000030_300 |
0.0973242394 |
- |
- |
Thioredoxin, putative [Ricinus communis] |
| 8 |
Hb_000926_280 |
0.0981711936 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 9 |
Hb_000061_370 |
0.0987966971 |
- |
- |
PREDICTED: zinc finger RNA-binding protein [Vitis vinifera] |
| 10 |
Hb_000579_140 |
0.0999168136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001677_210 |
0.1005643211 |
- |
- |
hypothetical protein JCGZ_01374 [Jatropha curcas] |
| 12 |
Hb_000629_010 |
0.1016529681 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 32 isoform X1 [Jatropha curcas] |
| 13 |
Hb_032208_090 |
0.1031497914 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 14 |
Hb_006827_020 |
0.1053448042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002530_030 |
0.1064297656 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 4 [Jatropha curcas] |
| 16 |
Hb_004046_050 |
0.1064905185 |
- |
- |
PREDICTED: origin of replication complex subunit 3 [Jatropha curcas] |
| 17 |
Hb_001758_100 |
0.1067665477 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000016_250 |
0.1076180398 |
- |
- |
unnamed protein product [Malassezia sympodialis ATCC 42132] |
| 19 |
Hb_000010_400 |
0.1078578034 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 67 [Jatropha curcas] |
| 20 |
Hb_000504_140 |
0.1078669158 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 26, chloroplastic-like [Jatropha curcas] |