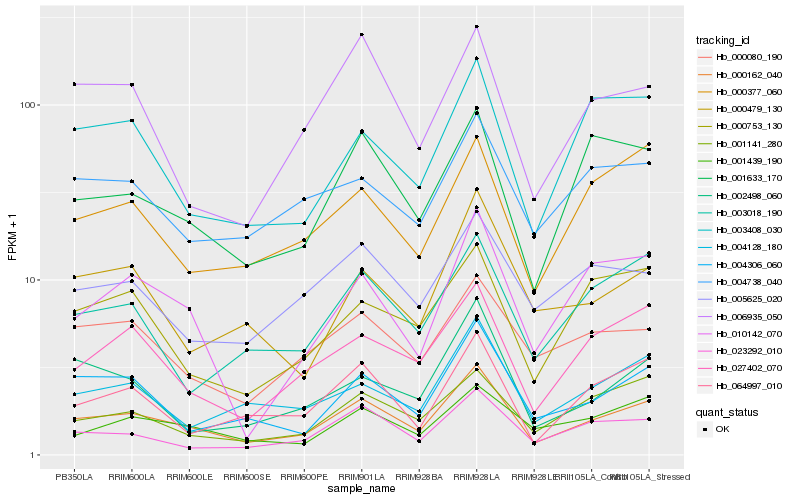
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000162_040 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 2 |
Hb_027402_070 |
0.1193830336 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 3 |
Hb_004306_060 |
0.1217610006 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g25060, mitochondrial [Jatropha curcas] |
| 4 |
Hb_023292_010 |
0.1262633894 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 5 |
Hb_004128_180 |
0.1270567376 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_064997_010 |
0.1292092351 |
- |
- |
hypothetical protein JCGZ_19852 [Jatropha curcas] |
| 7 |
Hb_001633_170 |
0.1302085027 |
- |
- |
PREDICTED: aspartokinase 2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001141_280 |
0.1346291597 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14820 [Vitis vinifera] |
| 9 |
Hb_010142_070 |
0.1346416545 |
- |
- |
PREDICTED: uncharacterized protein LOC105630659 [Jatropha curcas] |
| 10 |
Hb_000377_060 |
0.1351533476 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 11 |
Hb_000080_190 |
0.1352602398 |
- |
- |
- |
| 12 |
Hb_003018_190 |
0.1358872953 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g31070, mitochondrial [Jatropha curcas] |
| 13 |
Hb_003408_030 |
0.1369275362 |
- |
- |
PREDICTED: protein YIF1B [Jatropha curcas] |
| 14 |
Hb_006935_050 |
0.1385283807 |
- |
- |
PREDICTED: probable glutathione peroxidase 2 [Populus euphratica] |
| 15 |
Hb_001439_190 |
0.1392514594 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 16 |
Hb_004738_040 |
0.1399691714 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like isoform X2 [Populus euphratica] |
| 17 |
Hb_002498_060 |
0.1400547249 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_000753_130 |
0.1405541884 |
- |
- |
PREDICTED: uncharacterized protein LOC105640673 [Jatropha curcas] |
| 19 |
Hb_000479_130 |
0.1410280704 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39350 [Jatropha curcas] |
| 20 |
Hb_005625_020 |
0.1410974121 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 2 isoform X1 [Jatropha curcas] |