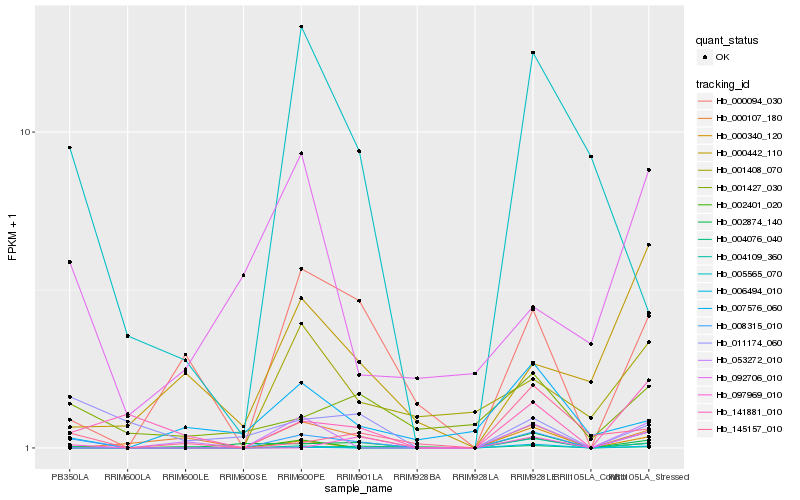
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008315_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105772042 [Gossypium raimondii] |
| 2 |
Hb_002401_020 |
0.3512699448 |
- |
- |
PREDICTED: protein argonaute 16 isoform X2 [Jatropha curcas] |
| 3 |
Hb_141881_010 |
0.3600551485 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_000094_030 |
0.3710965758 |
- |
- |
- |
| 5 |
Hb_001427_030 |
0.3920194586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000107_180 |
0.3978787357 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103343662 [Prunus mume] |
| 7 |
Hb_097969_010 |
0.3979436359 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_000340_120 |
0.3990760647 |
- |
- |
PREDICTED: uncharacterized protein LOC103406091 [Malus domestica] |
| 9 |
Hb_002874_140 |
0.4054051699 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 11 homolog [Prunus mume] |
| 10 |
Hb_011174_060 |
0.4078499307 |
- |
- |
- |
| 11 |
Hb_005565_070 |
0.4091559717 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_053272_010 |
0.412062968 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 13 |
Hb_145157_010 |
0.4140082124 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 14 |
Hb_004076_040 |
0.419561259 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_004109_360 |
0.4219547448 |
- |
- |
PREDICTED: omega-6 fatty acid desaturase, endoplasmic reticulum isozyme 1-like [Jatropha curcas] |
| 16 |
Hb_006494_010 |
0.425542499 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 17 |
Hb_007576_060 |
0.4269547553 |
- |
- |
- |
| 18 |
Hb_092706_010 |
0.4282756492 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 19 |
Hb_000442_110 |
0.4285046399 |
- |
- |
hypothetical protein POPTR_0010s18960g [Populus trichocarpa] |
| 20 |
Hb_001408_070 |
0.4300645826 |
- |
- |
- |