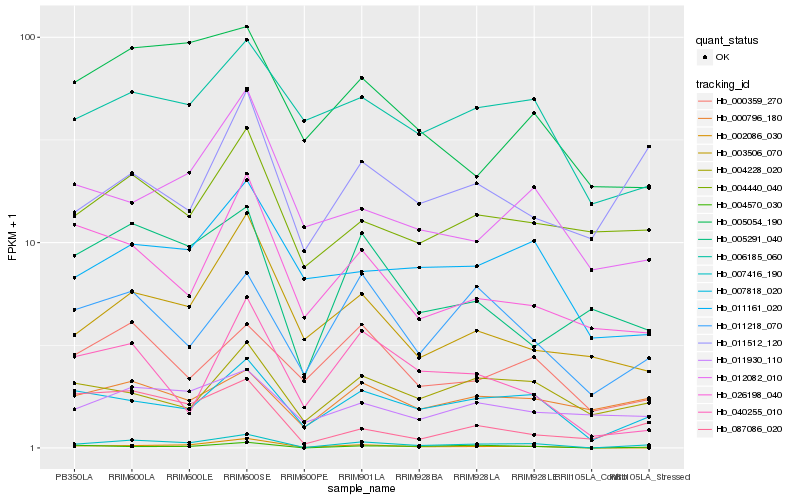
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007416_190 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 2 |
Hb_007818_020 |
0.1743942014 |
- |
- |
predicted protein [Physcomitrella patens] |
| 3 |
Hb_004570_030 |
0.1918926729 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011218_070 |
0.2164006842 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 5 |
Hb_003506_070 |
0.2171040231 |
- |
- |
PREDICTED: calcineurin B-like protein 7 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002086_030 |
0.2192561314 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 7 |
Hb_005291_040 |
0.2220277619 |
- |
- |
PREDICTED: gamma-gliadin [Jatropha curcas] |
| 8 |
Hb_026198_040 |
0.2255249862 |
- |
- |
PREDICTED: uncharacterized protein LOC105628901 [Jatropha curcas] |
| 9 |
Hb_040255_010 |
0.225758775 |
- |
- |
hypothetical protein JCGZ_08656 [Jatropha curcas] |
| 10 |
Hb_004440_040 |
0.225914839 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_011930_110 |
0.2264356768 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g59720, mitochondrial [Jatropha curcas] |
| 12 |
Hb_011161_020 |
0.2266879851 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 13 |
Hb_005054_190 |
0.2272469467 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000359_270 |
0.2274084055 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_004228_020 |
0.2275591268 |
- |
- |
hypothetical protein JCGZ_21866 [Jatropha curcas] |
| 16 |
Hb_012082_010 |
0.2286044129 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_006185_060 |
0.2290004557 |
- |
- |
PREDICTED: protein phosphatase 2C 16 [Jatropha curcas] |
| 18 |
Hb_087086_020 |
0.2295030142 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 19 |
Hb_000796_180 |
0.2296411687 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Vitis vinifera] |
| 20 |
Hb_011512_120 |
0.2298688971 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 1 [Jatropha curcas] |