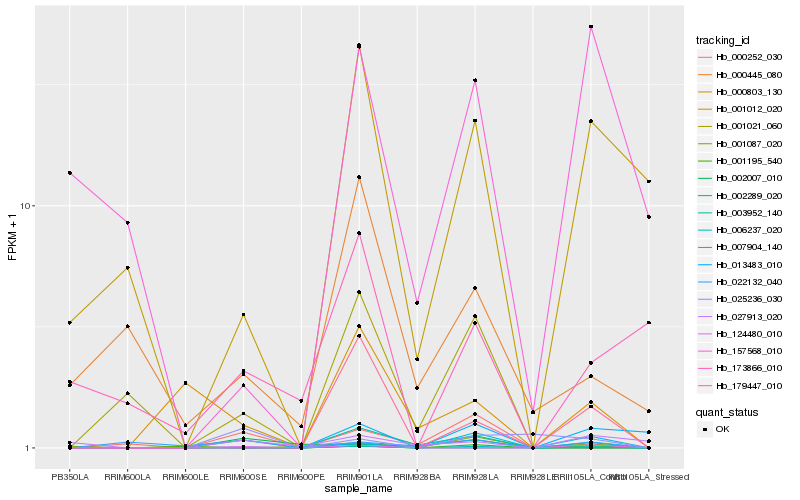
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006237_020 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 2 |
Hb_001012_020 |
0.3619824715 |
transcription factor |
TF Family: M-type |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_003952_140 |
0.3680283342 |
- |
- |
hypothetical protein POPTR_0001s24960g [Populus trichocarpa] |
| 4 |
Hb_179447_010 |
0.3744174951 |
- |
- |
hypothetical protein JCGZ_15340 [Jatropha curcas] |
| 5 |
Hb_007904_140 |
0.3847358986 |
- |
- |
- |
| 6 |
Hb_002007_010 |
0.3999857493 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 37a [Jatropha curcas] |
| 7 |
Hb_002289_020 |
0.4060202004 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 8 |
Hb_013483_010 |
0.4073739989 |
- |
- |
- |
| 9 |
Hb_000803_130 |
0.4082068193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000252_030 |
0.4105841021 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 11 |
Hb_022132_040 |
0.4113714541 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 12 |
Hb_001021_060 |
0.4209395015 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_157568_010 |
0.4240600342 |
- |
- |
MATE efflux family protein, putative [Theobroma cacao] |
| 14 |
Hb_001195_540 |
0.4253317463 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 15 |
Hb_124480_010 |
0.4274361365 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 16 |
Hb_025236_030 |
0.4279856737 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 17 |
Hb_173866_010 |
0.4322744562 |
- |
- |
- |
| 18 |
Hb_001087_020 |
0.4332388396 |
- |
- |
PREDICTED: phytoene synthase 2, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_027913_020 |
0.4346629375 |
- |
- |
- |
| 20 |
Hb_000445_080 |
0.4387565639 |
transcription factor |
TF Family: MIKC |
PREDICTED: floral homeotic protein PMADS 2 [Jatropha curcas] |