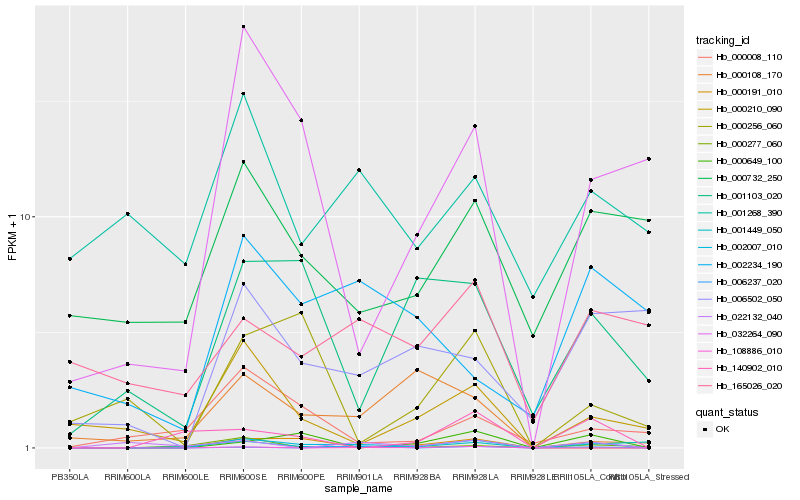
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002007_010 |
0.0 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 37a [Jatropha curcas] |
| 2 |
Hb_000649_100 |
0.352598212 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_032264_090 |
0.3597158596 |
- |
- |
hypothetical protein JCGZ_02512 [Jatropha curcas] |
| 4 |
Hb_001103_020 |
0.3604721287 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 5 |
Hb_000210_090 |
0.3811059641 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 6 |
Hb_140902_010 |
0.3894244865 |
- |
- |
PREDICTED: uncharacterized protein LOC105800955 [Gossypium raimondii] |
| 7 |
Hb_001449_050 |
0.3918659068 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000191_010 |
0.3928220472 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_002234_190 |
0.3993483016 |
- |
- |
PREDICTED: putative non-specific lipid-transfer protein 14 [Jatropha curcas] |
| 10 |
Hb_006237_020 |
0.3999857493 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 11 |
Hb_000732_250 |
0.400252364 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B2, mitochondrial-like [Populus euphratica] |
| 12 |
Hb_006502_050 |
0.4007062735 |
- |
- |
- |
| 13 |
Hb_000277_060 |
0.4027269198 |
- |
- |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 14 |
Hb_000008_110 |
0.4041390956 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Gossypium raimondii] |
| 15 |
Hb_001268_390 |
0.4048560615 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 16 |
Hb_000256_060 |
0.4072531493 |
- |
- |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 17 |
Hb_108886_010 |
0.4084169448 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_022132_040 |
0.4102634744 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 19 |
Hb_000108_170 |
0.4109344138 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 20 |
Hb_165026_020 |
0.4110846616 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0014s10300g [Populus trichocarpa] |