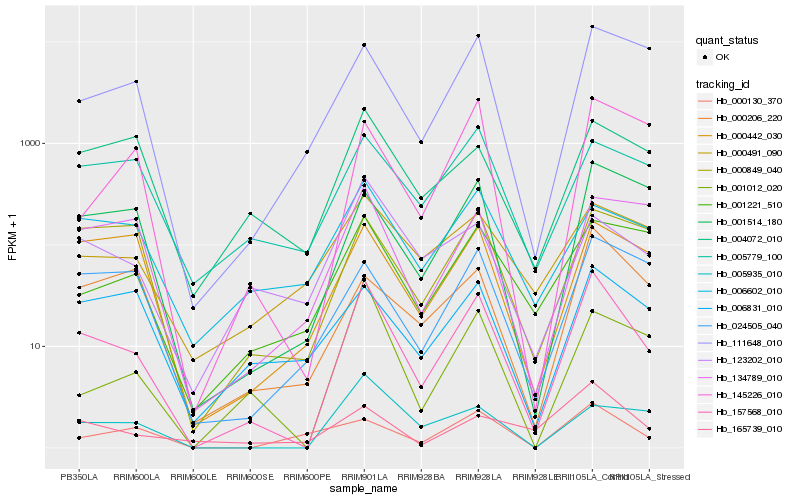
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_157568_010 |
0.0 |
- |
- |
MATE efflux family protein, putative [Theobroma cacao] |
| 2 |
Hb_111648_010 |
0.2176902788 |
rubber biosynthesis |
Gene Name: REF1 |
rubber elongation factor [Hevea brasiliensis] |
| 3 |
Hb_001514_180 |
0.218660204 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_16067 [Jatropha curcas] |
| 4 |
Hb_001012_020 |
0.2229610207 |
transcription factor |
TF Family: M-type |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_000442_030 |
0.2336311218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_024505_040 |
0.2342781447 |
- |
- |
homogentisate geranylgeranyl transferase [Hevea brasiliensis] |
| 7 |
Hb_001221_510 |
0.2380086265 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 8 |
Hb_145226_010 |
0.2385505904 |
- |
- |
PREDICTED: uncharacterized protein LOC100262861 [Vitis vinifera] |
| 9 |
Hb_000206_220 |
0.2395786572 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 9 [Jatropha curcas] |
| 10 |
Hb_134789_010 |
0.24018125 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 11 |
Hb_165739_010 |
0.2413506124 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_000849_040 |
0.2415820022 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_005779_100 |
0.2428360511 |
- |
- |
DNAJ [Theobroma cacao] |
| 14 |
Hb_004072_010 |
0.2444502212 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000491_090 |
0.2445922663 |
- |
- |
hypothetical protein B456_008G219400 [Gossypium raimondii] |
| 16 |
Hb_000130_370 |
0.2454014675 |
- |
- |
PREDICTED: uncharacterized protein LOC105635737 [Jatropha curcas] |
| 17 |
Hb_005935_010 |
0.2462905273 |
- |
- |
- |
| 18 |
Hb_006831_010 |
0.2464582322 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_006602_010 |
0.2465401363 |
- |
- |
PREDICTED: tobamovirus multiplication protein 3 [Jatropha curcas] |
| 20 |
Hb_123202_010 |
0.2482736856 |
- |
- |
thioredoxin h [Hevea brasiliensis] |