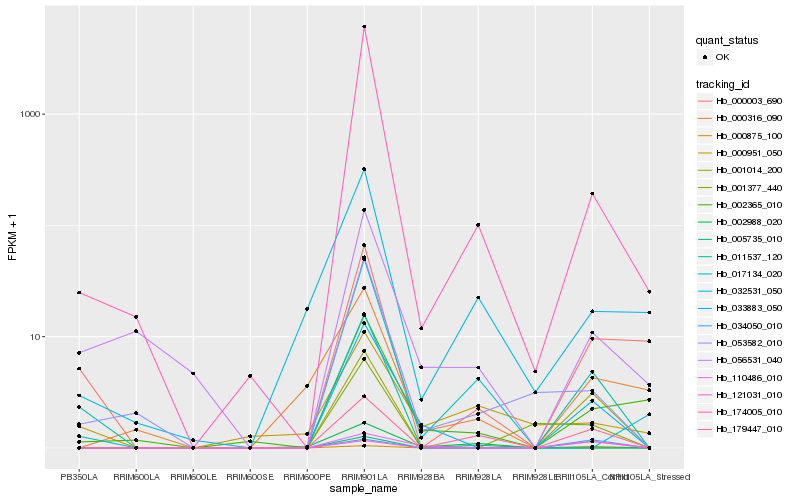
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_179447_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_15340 [Jatropha curcas] |
| 2 |
Hb_001014_200 |
0.2427029179 |
- |
- |
- |
| 3 |
Hb_033883_050 |
0.2924946512 |
- |
- |
- |
| 4 |
Hb_000875_100 |
0.2951804819 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 5 |
Hb_011537_120 |
0.297082002 |
- |
- |
- |
| 6 |
Hb_174005_010 |
0.3002384098 |
- |
- |
hypothetical protein EUGRSUZ_D00590 [Eucalyptus grandis] |
| 7 |
Hb_032531_050 |
0.3049807235 |
- |
- |
hypothetical protein CICLE_v10003980mg [Citrus clementina] |
| 8 |
Hb_005735_010 |
0.3149009194 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_110486_010 |
0.321740414 |
- |
- |
- |
| 10 |
Hb_053582_010 |
0.3235222812 |
- |
- |
- |
| 11 |
Hb_000003_690 |
0.3304553282 |
- |
- |
- |
| 12 |
Hb_121031_010 |
0.3327009734 |
- |
- |
PREDICTED: uncharacterized protein LOC104104926 [Nicotiana tomentosiformis] |
| 13 |
Hb_000316_090 |
0.3358373941 |
- |
- |
- |
| 14 |
Hb_034050_010 |
0.3397562498 |
- |
- |
PREDICTED: uncharacterized protein LOC103498182 [Cucumis melo] |
| 15 |
Hb_002365_010 |
0.3398492903 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 16 |
Hb_001377_440 |
0.3430233882 |
- |
- |
- |
| 17 |
Hb_017134_020 |
0.3445307108 |
- |
- |
- |
| 18 |
Hb_002988_020 |
0.3483804441 |
- |
- |
PREDICTED: uncharacterized protein LOC105637371 [Jatropha curcas] |
| 19 |
Hb_000951_050 |
0.3488620036 |
- |
- |
- |
| 20 |
Hb_056531_040 |
0.3507903451 |
- |
- |
PREDICTED: uncharacterized protein LOC103494239 [Cucumis melo] |