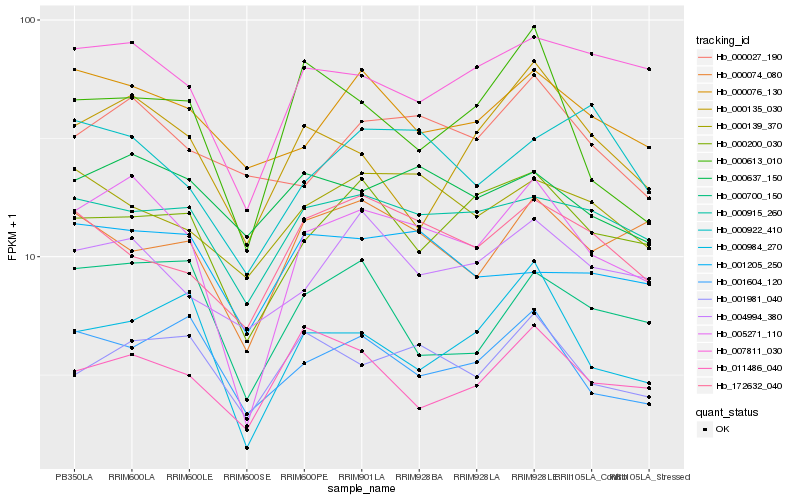
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005271_110 |
0.0 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 2 |
Hb_007811_030 |
0.130495618 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_000200_030 |
0.1359163599 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000984_270 |
0.1397647725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000915_260 |
0.1401120702 |
- |
- |
PREDICTED: uncharacterized protein LOC105628514 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001981_040 |
0.1417176366 |
transcription factor |
TF Family: PHD |
PREDICTED: origin of replication complex subunit 1B-like [Jatropha curcas] |
| 7 |
Hb_004994_380 |
0.1430788304 |
- |
- |
hypothetical protein JCGZ_14410 [Jatropha curcas] |
| 8 |
Hb_000700_150 |
0.1459589346 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000076_130 |
0.1466751598 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 10 |
Hb_001604_120 |
0.1486136515 |
- |
- |
PREDICTED: hemK methyltransferase family member 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000637_150 |
0.1498456362 |
- |
- |
Protein CASP, putative [Ricinus communis] |
| 12 |
Hb_000135_030 |
0.1501204372 |
- |
- |
PREDICTED: uncharacterized protein LOC105634729 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000139_370 |
0.1513888564 |
- |
- |
ubiquitin-activating enzyme E1c, putative [Ricinus communis] |
| 14 |
Hb_000074_080 |
0.1528517591 |
- |
- |
unknown [Populus trichocarpa] |
| 15 |
Hb_000027_190 |
0.152976959 |
- |
- |
PREDICTED: uncharacterized protein LOC105642057 isoform X1 [Jatropha curcas] |
| 16 |
Hb_172632_040 |
0.1534021794 |
- |
- |
PREDICTED: pyrophosphate-energized membrane proton pump 2 isoform X1 [Amborella trichopoda] |
| 17 |
Hb_011486_040 |
0.1534629077 |
- |
- |
PREDICTED: protein trichome birefringence-like 13 [Jatropha curcas] |
| 18 |
Hb_000613_010 |
0.1542312554 |
- |
- |
PREDICTED: uncharacterized protein LOC105647991 [Jatropha curcas] |
| 19 |
Hb_001205_250 |
0.1575506586 |
- |
- |
PREDICTED: uncharacterized protein LOC105643856 [Jatropha curcas] |
| 20 |
Hb_000922_410 |
0.1588296678 |
- |
- |
PREDICTED: anthranilate phosphoribosyltransferase, chloroplastic [Jatropha curcas] |