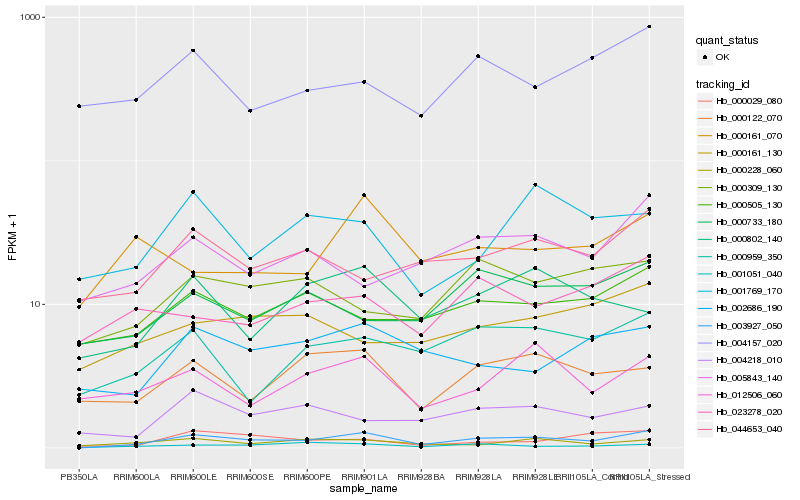
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003927_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629028 [Jatropha curcas] |
| 2 |
Hb_000959_350 |
0.2109688796 |
- |
- |
PREDICTED: uncharacterized protein LOC105636607 [Jatropha curcas] |
| 3 |
Hb_000122_070 |
0.2125021952 |
- |
- |
hypothetical protein POPTR_0010s18960g [Populus trichocarpa] |
| 4 |
Hb_002686_190 |
0.2147904876 |
- |
- |
- |
| 5 |
Hb_001051_040 |
0.2161254516 |
- |
- |
PREDICTED: uncharacterized protein LOC105628076 [Jatropha curcas] |
| 6 |
Hb_000228_060 |
0.2198217629 |
- |
- |
hypothetical protein JCGZ_21532 [Jatropha curcas] |
| 7 |
Hb_000029_080 |
0.2199405427 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein B-like [Jatropha curcas] |
| 8 |
Hb_012506_060 |
0.2202391483 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 9 |
Hb_000505_130 |
0.2221031515 |
- |
- |
PREDICTED: uncharacterized protein LOC105641262 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000802_140 |
0.2237374438 |
- |
- |
hypothetical protein B456_011G045700 [Gossypium raimondii] |
| 11 |
Hb_004218_010 |
0.2243338811 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 12 |
Hb_044653_040 |
0.2245111733 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 13 |
Hb_000309_130 |
0.2277117845 |
- |
- |
hypothetical protein JCGZ_09592 [Jatropha curcas] |
| 14 |
Hb_004157_020 |
0.2287850647 |
- |
- |
PREDICTED: histone H2B-like [Gossypium raimondii] |
| 15 |
Hb_001769_170 |
0.2293113997 |
- |
- |
50S ribosomal protein L31, putative [Ricinus communis] |
| 16 |
Hb_023278_020 |
0.2317478812 |
- |
- |
Peroxin 3 isoform 1 [Theobroma cacao] |
| 17 |
Hb_000733_180 |
0.2320106229 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein B-like [Jatropha curcas] |
| 18 |
Hb_005843_140 |
0.2328707415 |
- |
- |
Red chlorophyll catabolite reductase, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000161_130 |
0.2332807624 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000161_070 |
0.2344462665 |
- |
- |
PREDICTED: ATP phosphoribosyltransferase 2, chloroplastic-like [Jatropha curcas] |