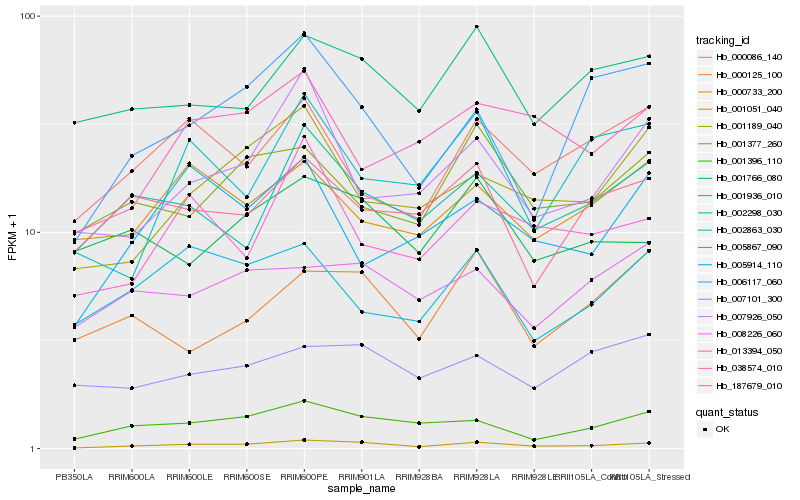
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001051_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628076 [Jatropha curcas] |
| 2 |
Hb_001396_110 |
0.1329174096 |
- |
- |
DNA primase large subunit, putative [Ricinus communis] |
| 3 |
Hb_006117_060 |
0.1475865833 |
- |
- |
hypothetical protein CARUB_v10024452mg [Capsella rubella] |
| 4 |
Hb_001936_010 |
0.1517667015 |
- |
- |
hypothetical protein Csa_2G348280 [Cucumis sativus] |
| 5 |
Hb_001189_040 |
0.1542871372 |
- |
- |
hypothetical protein POPTR_0014s10420g [Populus trichocarpa] |
| 6 |
Hb_002298_030 |
0.154467111 |
- |
- |
PREDICTED: coenzyme Q-binding protein COQ10 homolog, mitochondrial [Jatropha curcas] |
| 7 |
Hb_007926_050 |
0.1619029001 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 8 |
Hb_013394_050 |
0.1648332009 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 [Nicotiana sylvestris] |
| 9 |
Hb_008226_060 |
0.1659841238 |
- |
- |
PREDICTED: folic acid synthesis protein fol1-like [Jatropha curcas] |
| 10 |
Hb_001377_260 |
0.1670153298 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_000086_140 |
0.1671639462 |
- |
- |
PREDICTED: probable arabinose 5-phosphate isomerase [Jatropha curcas] |
| 12 |
Hb_000125_100 |
0.1672018336 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105635070 [Jatropha curcas] |
| 13 |
Hb_038574_010 |
0.1681779515 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like isoform X2 [Populus euphratica] |
| 14 |
Hb_002863_030 |
0.168977539 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 15 |
Hb_001766_080 |
0.1692243209 |
- |
- |
PREDICTED: uncharacterized protein LOC105644125 [Jatropha curcas] |
| 16 |
Hb_007101_300 |
0.1697871567 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial [Jatropha curcas] |
| 17 |
Hb_005867_090 |
0.1703553487 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 18 |
Hb_187679_010 |
0.1707510052 |
- |
- |
- |
| 19 |
Hb_005914_110 |
0.1714453521 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 20 |
Hb_000733_200 |
0.1724060441 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |