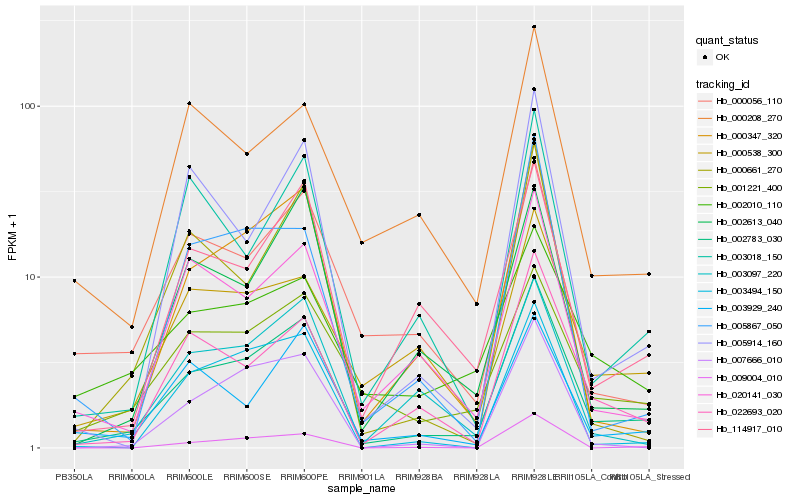
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002783_030 |
0.0 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 2 |
Hb_000347_320 |
0.1429155857 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 3 |
Hb_022693_020 |
0.146385592 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 4 |
Hb_114917_010 |
0.1530072834 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001221_400 |
0.1579282129 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 6 |
Hb_020141_030 |
0.1614184095 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 7 |
Hb_003097_220 |
0.1615423729 |
- |
- |
hypothetical protein POPTR_0001s06150g [Populus trichocarpa] |
| 8 |
Hb_000538_300 |
0.1689669453 |
- |
- |
PREDICTED: WAT1-related protein At3g02690, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003929_240 |
0.1697875633 |
- |
- |
PREDICTED: uncharacterized protein LOC105643616 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002613_040 |
0.1698276206 |
- |
- |
phosphate translocator-related family protein [Populus trichocarpa] |
| 11 |
Hb_003018_150 |
0.1699966719 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 12 |
Hb_000056_110 |
0.1726893723 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 13 |
Hb_003494_150 |
0.1766806147 |
- |
- |
Silicon transporter, putative [Ricinus communis] |
| 14 |
Hb_005914_160 |
0.1774466082 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000661_270 |
0.1777830731 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_009004_010 |
0.1794946035 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 17 |
Hb_007666_010 |
0.1801634089 |
- |
- |
PREDICTED: uncharacterized protein LOC105649266 [Jatropha curcas] |
| 18 |
Hb_000208_270 |
0.1809516465 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 19 |
Hb_002010_110 |
0.1810156063 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 20 |
Hb_005867_050 |
0.1833743099 |
- |
- |
PREDICTED: protein ECERIFERUM 1 [Jatropha curcas] |