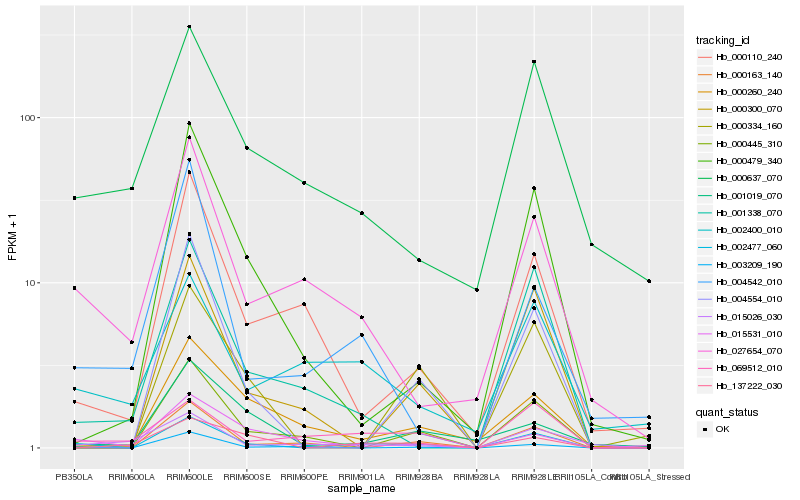
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002477_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004554_010 |
0.2611465843 |
- |
- |
PREDICTED: uncharacterized protein LOC105636233 isoform X1 [Jatropha curcas] |
| 3 |
Hb_027654_070 |
0.2735916436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_069512_010 |
0.2833015877 |
- |
- |
- |
| 5 |
Hb_000300_070 |
0.2840020883 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 6 |
Hb_003209_190 |
0.2859286454 |
- |
- |
hypothetical protein POPTR_0010s16770g [Populus trichocarpa] |
| 7 |
Hb_000479_340 |
0.2871771813 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 8 |
Hb_000260_240 |
0.2879144101 |
- |
- |
hypothetical protein JCGZ_00622 [Jatropha curcas] |
| 9 |
Hb_001338_070 |
0.2886819779 |
- |
- |
ORF126 [Jatropha curcas] |
| 10 |
Hb_000334_160 |
0.2897004205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000445_310 |
0.2898875048 |
- |
- |
hypothetical protein B456_011G218000 [Gossypium raimondii] |
| 12 |
Hb_137222_030 |
0.2927981281 |
- |
- |
- |
| 13 |
Hb_000637_070 |
0.29390893 |
- |
- |
PREDICTED: uncharacterized protein LOC105650668 [Jatropha curcas] |
| 14 |
Hb_004542_010 |
0.2939268011 |
- |
- |
Putative chloroplast RF19 [Medicago truncatula] |
| 15 |
Hb_000110_240 |
0.2967049985 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 16 |
Hb_015026_030 |
0.2991172717 |
- |
- |
hypothetical protein POPTR_0011s04350g [Populus trichocarpa] |
| 17 |
Hb_015531_010 |
0.3007312196 |
- |
- |
PREDICTED: uncharacterized protein LOC105636764 [Jatropha curcas] |
| 18 |
Hb_000163_140 |
0.3014769877 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 19 |
Hb_001019_070 |
0.3017436977 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 20 |
Hb_002400_010 |
0.3018730526 |
- |
- |
PREDICTED: putative L-ascorbate peroxidase 6 [Jatropha curcas] |