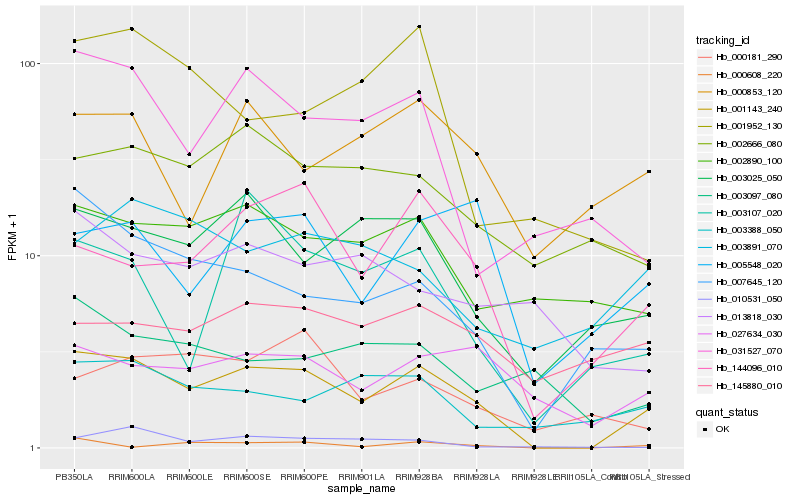
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001143_240 |
0.0 |
- |
- |
8-oxoguanine DNA glycosylase, putative [Ricinus communis] |
| 2 |
Hb_144096_010 |
0.2075755226 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 3 |
Hb_007645_120 |
0.2111631326 |
- |
- |
hypothetical protein POPTR_0006s05370g [Populus trichocarpa] |
| 4 |
Hb_003025_050 |
0.2122966355 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 5 |
Hb_005548_020 |
0.2253084716 |
- |
- |
hypothetical protein JCGZ_05605 [Jatropha curcas] |
| 6 |
Hb_031527_070 |
0.2276100412 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 7 |
Hb_027634_030 |
0.228959324 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_002890_100 |
0.2322975723 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein MAD1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002666_080 |
0.2334142909 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_003388_050 |
0.2354218391 |
- |
- |
- |
| 11 |
Hb_000853_120 |
0.2416275654 |
- |
- |
PREDICTED: ethylene response sensor 1 [Jatropha curcas] |
| 12 |
Hb_003107_020 |
0.2427265222 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 13 |
Hb_003891_070 |
0.2459264874 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 14 |
Hb_000181_290 |
0.2485288234 |
- |
- |
hypothetical protein POPTR_0001s41000g [Populus trichocarpa] |
| 15 |
Hb_010531_050 |
0.2505130193 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001952_130 |
0.2515219154 |
- |
- |
PREDICTED: protein EXORDIUM-like 2 [Jatropha curcas] |
| 17 |
Hb_145880_010 |
0.2516506645 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 18 |
Hb_000608_220 |
0.2522021867 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 19 |
Hb_003097_080 |
0.253633327 |
- |
- |
hypothetical protein JCGZ_24481 [Jatropha curcas] |
| 20 |
Hb_013818_030 |
0.2537716862 |
- |
- |
conserved hypothetical protein [Ricinus communis] |