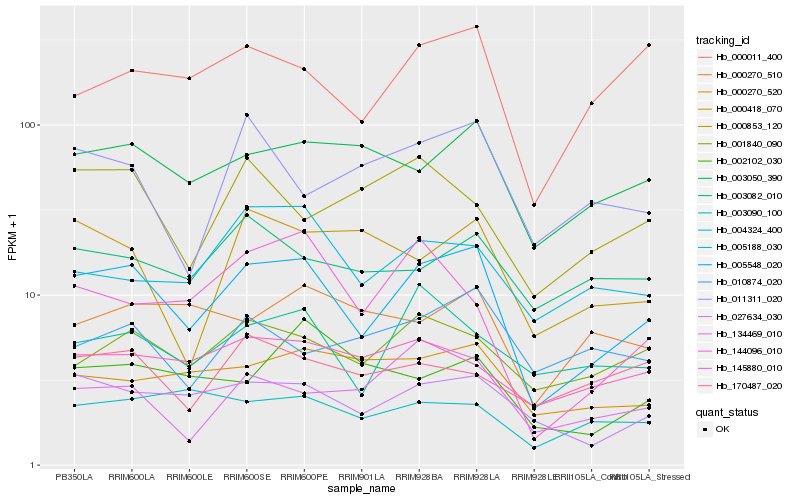
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005548_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_05605 [Jatropha curcas] |
| 2 |
Hb_027634_030 |
0.1451979004 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_003050_390 |
0.1611436582 |
- |
- |
PREDICTED: uncharacterized protein LOC104442543 [Eucalyptus grandis] |
| 4 |
Hb_001840_090 |
0.1649655592 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 5 |
Hb_000011_400 |
0.1698773172 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 4 [Hevea brasiliensis] |
| 6 |
Hb_003090_100 |
0.1760262183 |
- |
- |
PREDICTED: uncharacterized protein LOC105646670 [Jatropha curcas] |
| 7 |
Hb_000270_520 |
0.1775828833 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_134469_010 |
0.1791456069 |
- |
- |
PREDICTED: protein argonaute 5 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005188_030 |
0.1813248884 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_145880_010 |
0.1822659241 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 11 |
Hb_003082_010 |
0.184500879 |
- |
- |
PREDICTED: putative F-box protein At3g10240 [Jatropha curcas] |
| 12 |
Hb_004324_400 |
0.1878831179 |
- |
- |
PREDICTED: protein timeless homolog [Jatropha curcas] |
| 13 |
Hb_000270_510 |
0.1902307253 |
- |
- |
hypothetical protein CISIN_1g008173mg [Citrus sinensis] |
| 14 |
Hb_000418_070 |
0.1905415308 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 15 |
Hb_011311_020 |
0.1918956655 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000853_120 |
0.1927088158 |
- |
- |
PREDICTED: ethylene response sensor 1 [Jatropha curcas] |
| 17 |
Hb_144096_010 |
0.1934137558 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 18 |
Hb_002102_030 |
0.1937074993 |
- |
- |
- |
| 19 |
Hb_010874_020 |
0.1941666672 |
- |
- |
PREDICTED: uncharacterized protein LOC105636745 [Jatropha curcas] |
| 20 |
Hb_170487_020 |
0.1969274915 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |