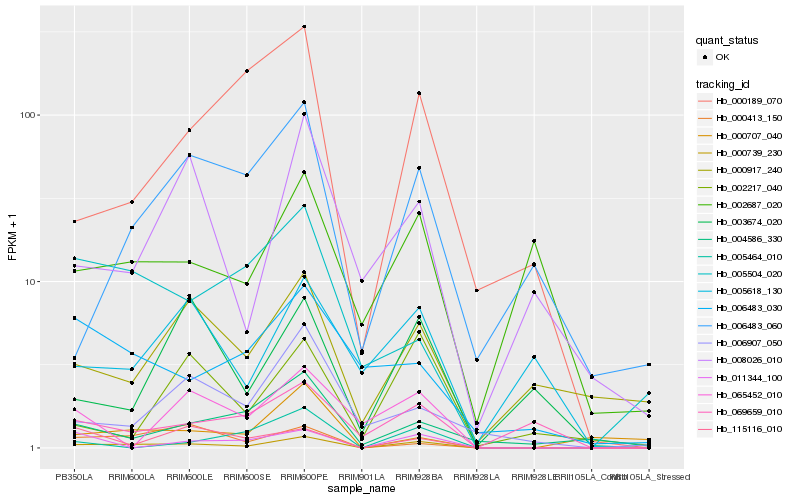
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000739_230 |
0.0 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 22 [Jatropha curcas] |
| 2 |
Hb_000413_150 |
0.2711784775 |
- |
- |
PREDICTED: uncharacterized protein LOC103709996 [Phoenix dactylifera] |
| 3 |
Hb_005504_020 |
0.2934838846 |
- |
- |
- |
| 4 |
Hb_069659_010 |
0.2953213692 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 5 |
Hb_000917_240 |
0.296481437 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 6 |
Hb_000189_070 |
0.2997973617 |
transcription factor |
TF Family: AUX/IAA |
Auxin-induced protein 22D, putative [Ricinus communis] |
| 7 |
Hb_115116_010 |
0.299865612 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 8 |
Hb_008026_010 |
0.3008511032 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 9 [Jatropha curcas] |
| 9 |
Hb_005618_130 |
0.3022694494 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 10 |
Hb_004586_330 |
0.3024027519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_011344_100 |
0.3024057399 |
- |
- |
- |
| 12 |
Hb_002687_020 |
0.3027715405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006907_050 |
0.3034271424 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_006483_060 |
0.3061898557 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 15 |
Hb_003674_020 |
0.3082833938 |
- |
- |
PREDICTED: DNA replication licensing factor MCM6 [Jatropha curcas] |
| 16 |
Hb_000707_040 |
0.3091045843 |
- |
- |
Uncharacterized protein TCM_021829 [Theobroma cacao] |
| 17 |
Hb_002217_040 |
0.3120889016 |
- |
- |
PREDICTED: boron transporter 1-like [Jatropha curcas] |
| 18 |
Hb_006483_030 |
0.3125200702 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 19 |
Hb_065452_010 |
0.3130845513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005464_010 |
0.3140831342 |
- |
- |
Thioredoxin H-type, putative [Ricinus communis] |