| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
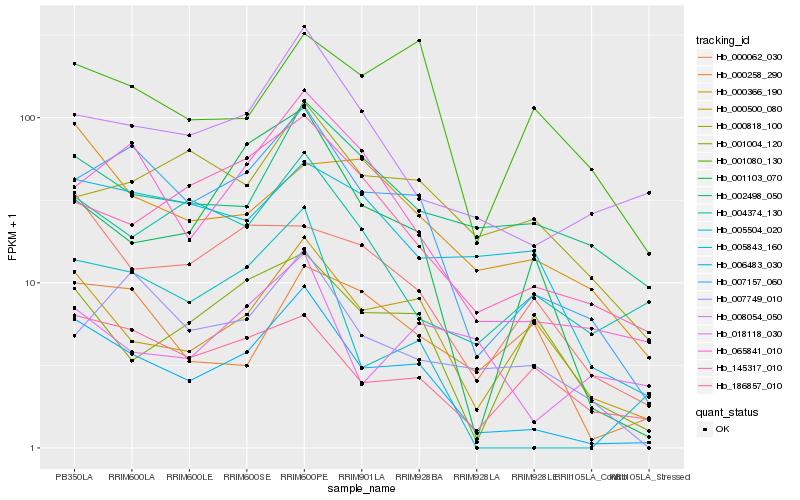
| 1 |
Hb_006483_030 |
0.0 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 2 |
Hb_007157_060 |
0.1455732511 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 3 |
Hb_005504_020 |
0.1905387196 |
- |
- |
- |
| 4 |
Hb_000500_080 |
0.1917883955 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 5 |
Hb_065841_010 |
0.193873052 |
- |
- |
hypothetical protein JCGZ_18376 [Jatropha curcas] |
| 6 |
Hb_001103_070 |
0.2134373834 |
- |
- |
Patellin-3, putative [Ricinus communis] |
| 7 |
Hb_008054_050 |
0.2141955492 |
- |
- |
PREDICTED: uncharacterized protein LOC101212042 [Cucumis sativus] |
| 8 |
Hb_145317_010 |
0.2197780454 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 9 |
Hb_000062_030 |
0.2250363642 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X2 [Jatropha curcas] |
| 10 |
Hb_004374_130 |
0.2290851917 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 11 |
Hb_005843_160 |
0.2298681197 |
- |
- |
PREDICTED: uncharacterized protein LOC105112024 [Populus euphratica] |
| 12 |
Hb_186857_010 |
0.2310481925 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 13 |
Hb_002498_050 |
0.2363787876 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |
| 14 |
Hb_001004_120 |
0.2395887549 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_007749_010 |
0.2478327162 |
- |
- |
- |
| 16 |
Hb_000366_190 |
0.2487969987 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 17 |
Hb_000818_100 |
0.2517603507 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |
| 18 |
Hb_018118_030 |
0.2546373034 |
- |
- |
PREDICTED: putative multidrug resistance protein, partial [Musa acuminata subsp. malaccensis] |
| 19 |
Hb_000258_290 |
0.2548463009 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP15 [Jatropha curcas] |
| 20 |
Hb_001080_130 |
0.2548948529 |
- |
- |
catalase [Hevea brasiliensis] |