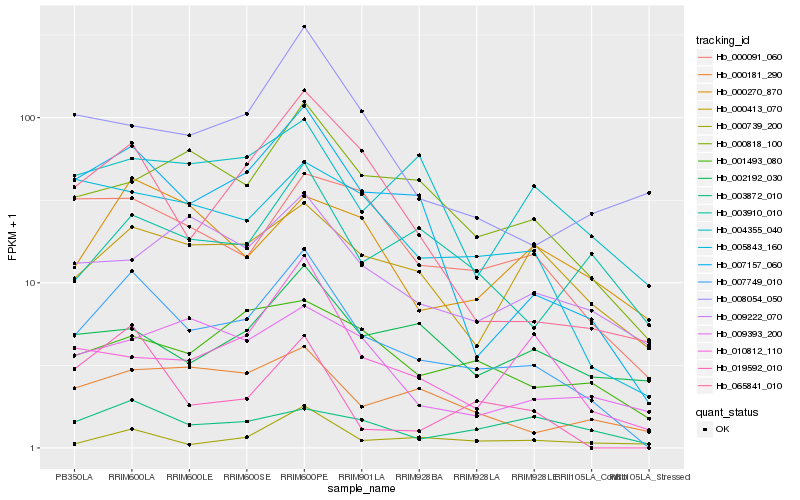
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007749_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_007157_060 |
0.1512849373 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 3 |
Hb_000818_100 |
0.1895545175 |
- |
- |
PREDICTED: delta(24)-sterol reductase [Jatropha curcas] |
| 4 |
Hb_065841_010 |
0.1909584471 |
- |
- |
hypothetical protein JCGZ_18376 [Jatropha curcas] |
| 5 |
Hb_005843_160 |
0.190991927 |
- |
- |
PREDICTED: uncharacterized protein LOC105112024 [Populus euphratica] |
| 6 |
Hb_000091_060 |
0.1992670315 |
- |
- |
PREDICTED: galactose oxidase [Jatropha curcas] |
| 7 |
Hb_000181_290 |
0.2011715559 |
- |
- |
hypothetical protein POPTR_0001s41000g [Populus trichocarpa] |
| 8 |
Hb_000413_070 |
0.2096687021 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001493_080 |
0.2114961181 |
- |
- |
PREDICTED: RING-H2 finger protein ATL3-like [Jatropha curcas] |
| 10 |
Hb_019592_010 |
0.2179320067 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 11 |
Hb_003910_010 |
0.2180771298 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 12 |
Hb_009222_070 |
0.2205207056 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 13 |
Hb_010812_110 |
0.2211884643 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 14 |
Hb_003872_010 |
0.2217271017 |
- |
- |
hypothetical protein PRUPE_ppa013215mg [Prunus persica] |
| 15 |
Hb_008054_050 |
0.2248494944 |
- |
- |
PREDICTED: uncharacterized protein LOC101212042 [Cucumis sativus] |
| 16 |
Hb_002192_030 |
0.2256034156 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004355_040 |
0.2260364206 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 18 |
Hb_009393_200 |
0.2262178018 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 19 |
Hb_000739_200 |
0.2262330312 |
transcription factor |
TF Family: LOB |
hypothetical protein CICLE_v10021992mg [Citrus clementina] |
| 20 |
Hb_000270_870 |
0.2265401707 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |