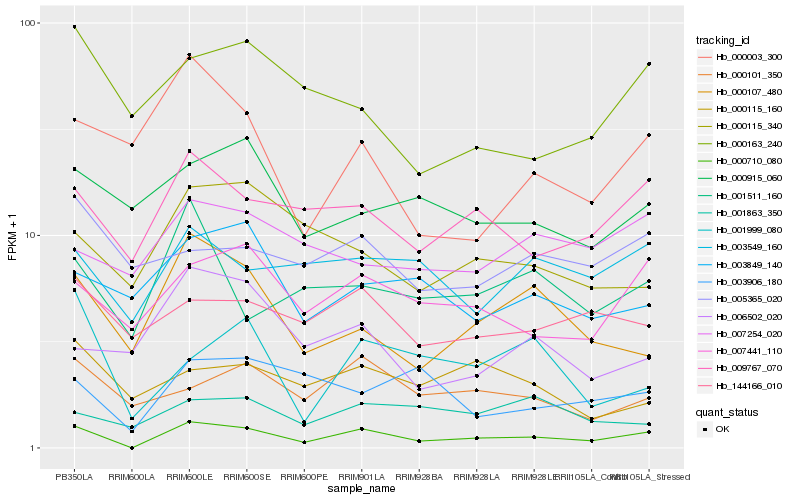
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000710_080 |
0.0 |
- |
- |
hypothetical protein EUTSA_v10023022mg [Eutrema salsugineum] |
| 2 |
Hb_009767_070 |
0.2109431572 |
- |
- |
PREDICTED: uncharacterized protein LOC105631980 [Jatropha curcas] |
| 3 |
Hb_007441_110 |
0.2148646681 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At3g58900-like [Jatropha curcas] |
| 4 |
Hb_000107_480 |
0.2235501586 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000101_350 |
0.2324375806 |
desease resistance |
Gene Name: NB-ARC |
NB-ARC domain-containing disease resistance protein, putative [Theobroma cacao] |
| 6 |
Hb_003549_160 |
0.2327691241 |
- |
- |
Protein FAM18B, putative [Ricinus communis] |
| 7 |
Hb_001511_160 |
0.2344752234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001999_080 |
0.2357612 |
- |
- |
CCCH-type zinc finger protein [Hevea brasiliensis] |
| 9 |
Hb_144166_010 |
0.2366646421 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 10 |
Hb_000003_300 |
0.2423453999 |
- |
- |
PREDICTED: uncharacterized protein LOC105631182 [Jatropha curcas] |
| 11 |
Hb_000115_340 |
0.245366852 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 12 |
Hb_000163_240 |
0.2454326738 |
- |
- |
PREDICTED: protein OPI10 homolog [Jatropha curcas] |
| 13 |
Hb_003849_140 |
0.2457671135 |
- |
- |
PREDICTED: uncharacterized protein LOC105644697 [Jatropha curcas] |
| 14 |
Hb_003906_180 |
0.2459543936 |
- |
- |
unknown [Zea mays] |
| 15 |
Hb_000915_060 |
0.2473966092 |
- |
- |
PREDICTED: uncharacterized protein LOC105628864 [Jatropha curcas] |
| 16 |
Hb_001863_350 |
0.2487602265 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 17 |
Hb_005365_020 |
0.2506430744 |
- |
- |
hypothetical protein JCGZ_15105 [Jatropha curcas] |
| 18 |
Hb_007254_020 |
0.2506948051 |
- |
- |
PREDICTED: uncharacterized protein LOC105629137 [Jatropha curcas] |
| 19 |
Hb_006502_020 |
0.25093012 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Jatropha curcas] |
| 20 |
Hb_000115_160 |
0.2520685682 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |