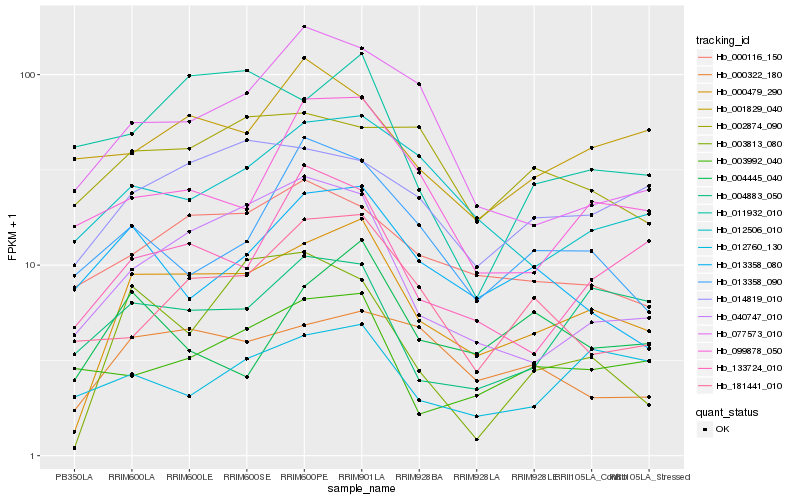
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000479_290 |
0.0 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000322_180 |
0.1725271938 |
- |
- |
recA family protein [Populus trichocarpa] |
| 3 |
Hb_040747_010 |
0.1785397448 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 4 |
Hb_012506_010 |
0.1789804611 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 5 |
Hb_014819_010 |
0.1798484475 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000116_150 |
0.1810949079 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 7 |
Hb_133724_010 |
0.1848096697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_181441_010 |
0.1921973526 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 9 |
Hb_004883_050 |
0.1928122197 |
- |
- |
PREDICTED: probable UDP-arabinopyranose mutase 5 isoform X2 [Vitis vinifera] |
| 10 |
Hb_099878_050 |
0.1931478912 |
- |
- |
PREDICTED: uncharacterized protein LOC105123495 isoform X2 [Populus euphratica] |
| 11 |
Hb_077573_010 |
0.2024572626 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_013358_080 |
0.2032564368 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 13 |
Hb_003813_080 |
0.2037248888 |
- |
- |
PREDICTED: uncharacterized protein LOC105133003 [Populus euphratica] |
| 14 |
Hb_003992_040 |
0.2042367405 |
- |
- |
Post-illumination chlorophyll fluorescence increase isoform 5 [Theobroma cacao] |
| 15 |
Hb_002874_090 |
0.2052501468 |
- |
- |
PREDICTED: uncharacterized protein LOC105637452 [Jatropha curcas] |
| 16 |
Hb_012760_130 |
0.2056974326 |
- |
- |
PREDICTED: uncharacterized protein LOC105647093 [Jatropha curcas] |
| 17 |
Hb_004445_040 |
0.2083403196 |
- |
- |
PREDICTED: uncharacterized protein LOC105639134 isoform X5 [Jatropha curcas] |
| 18 |
Hb_013358_090 |
0.2083832108 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 19 |
Hb_001829_040 |
0.2092182899 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 20 |
Hb_011932_010 |
0.2096837157 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |