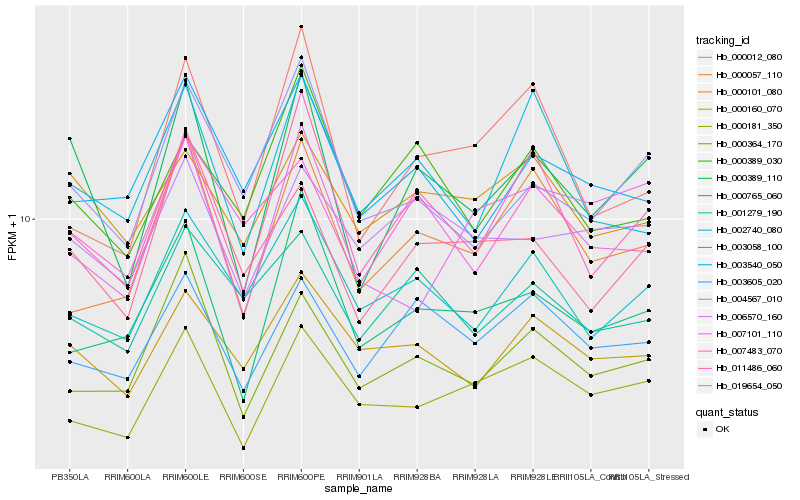
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000389_110 |
0.0 |
- |
- |
adenosine kinase, putative [Ricinus communis] |
| 2 |
Hb_011486_060 |
0.118047045 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004567_010 |
0.1386611912 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 4 |
Hb_019654_050 |
0.1399296954 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 5 |
Hb_000765_060 |
0.149039528 |
- |
- |
Alcohol dehydrogenase-like 6 [Glycine soja] |
| 6 |
Hb_000364_170 |
0.150716245 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 7 |
Hb_003605_020 |
0.1523918673 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 8 |
Hb_000160_070 |
0.154375353 |
- |
- |
PREDICTED: uncharacterized protein LOC105648219 [Jatropha curcas] |
| 9 |
Hb_002740_080 |
0.1568294325 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 10 |
Hb_006570_160 |
0.1572679167 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 11 |
Hb_000101_080 |
0.1602361284 |
- |
- |
PREDICTED: protein CREG1 [Jatropha curcas] |
| 12 |
Hb_000181_350 |
0.1610227115 |
- |
- |
PREDICTED: uncharacterized protein LOC105111090 [Populus euphratica] |
| 13 |
Hb_003058_100 |
0.1622311721 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000057_110 |
0.1624176619 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 15 |
Hb_007483_070 |
0.1645570667 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 16 |
Hb_000389_030 |
0.1646037915 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 17 |
Hb_003540_050 |
0.1669921692 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 18 |
Hb_007101_110 |
0.1672926267 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 19 |
Hb_001279_190 |
0.1678138986 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000012_080 |
0.1692171555 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |