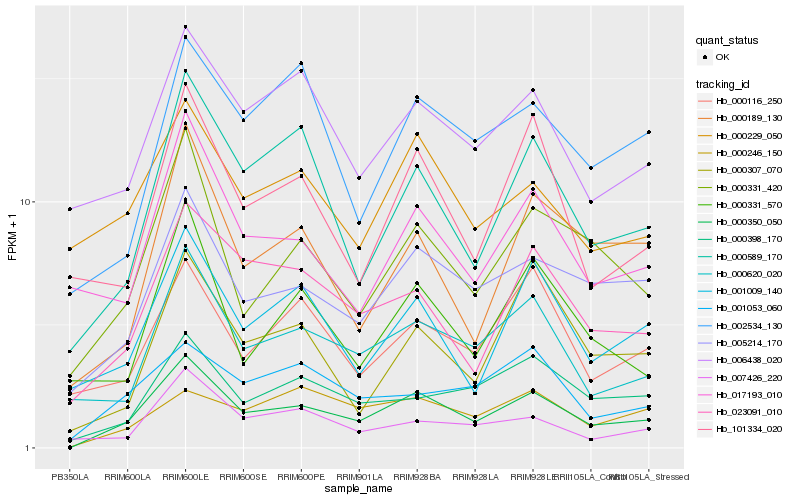
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000350_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 2 |
Hb_005214_170 |
0.1368251237 |
- |
- |
PREDICTED: uncharacterized protein LOC105636021 [Jatropha curcas] |
| 3 |
Hb_000589_170 |
0.1447955521 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000331_420 |
0.1467434003 |
- |
- |
PREDICTED: nucleoside diphosphate kinase 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_023091_010 |
0.1477179042 |
- |
- |
hypothetical protein POPTR_0016s06670g [Populus trichocarpa] |
| 6 |
Hb_000620_020 |
0.1499181847 |
- |
- |
PREDICTED: uncharacterized protein LOC105649917 [Jatropha curcas] |
| 7 |
Hb_000398_170 |
0.1529421878 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1-like [Jatropha curcas] |
| 8 |
Hb_001009_140 |
0.1609227737 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 9 |
Hb_000189_130 |
0.163380748 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001053_060 |
0.1642216705 |
- |
- |
DHHC-type zinc finger family protein [Theobroma cacao] |
| 11 |
Hb_006438_020 |
0.1665109857 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 12 |
Hb_000246_150 |
0.1675096195 |
- |
- |
PREDICTED: protein FAM135B [Jatropha curcas] |
| 13 |
Hb_101334_020 |
0.1681417957 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 14 |
Hb_017193_010 |
0.1686653985 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000116_250 |
0.1699171305 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 16 |
Hb_002534_130 |
0.1712253202 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 17 |
Hb_000307_070 |
0.1716959341 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 18 |
Hb_000331_570 |
0.1737491012 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase [Jatropha curcas] |
| 19 |
Hb_007426_220 |
0.1744701443 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000229_050 |
0.1752861199 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |