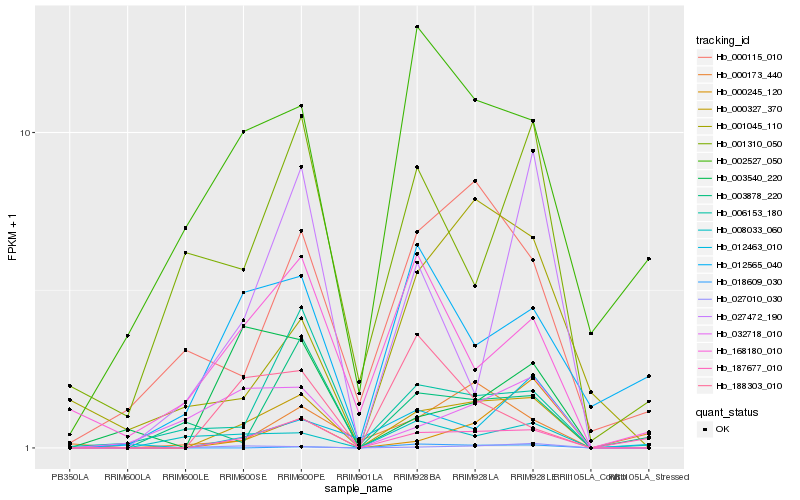
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000327_370 |
0.0 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 2 |
Hb_187677_010 |
0.2340269076 |
- |
- |
- |
| 3 |
Hb_027010_030 |
0.2792067433 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 4 |
Hb_006153_180 |
0.2902863083 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000245_120 |
0.2944218097 |
- |
- |
hypothetical protein POPTR_0001s18890g [Populus trichocarpa] |
| 6 |
Hb_000115_010 |
0.2957840739 |
- |
- |
5-methyltetrahydrofolate:homocysteine methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_032718_010 |
0.2969670944 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 8 |
Hb_008033_060 |
0.2970479329 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 9 |
Hb_027472_190 |
0.3072257943 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 10 |
Hb_001045_110 |
0.3148876039 |
transcription factor |
TF Family: M-type |
flowering locus C-like protein [Juglans regia] |
| 11 |
Hb_012463_010 |
0.3160971043 |
- |
- |
PREDICTED: uncharacterized protein LOC104901296 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_003540_220 |
0.3198456718 |
- |
- |
PREDICTED: thioredoxin [Jatropha curcas] |
| 13 |
Hb_012565_040 |
0.3223659546 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 14 |
Hb_018609_030 |
0.3237640363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_168180_010 |
0.3242384593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003878_220 |
0.3252924529 |
- |
- |
vacuolar protein sorting, putative [Ricinus communis] |
| 17 |
Hb_002527_050 |
0.3256743835 |
- |
- |
hypothetical protein JCGZ_20265 [Jatropha curcas] |
| 18 |
Hb_000173_440 |
0.3260148575 |
- |
- |
YALI0B05280p [Yarrowia lipolytica] |
| 19 |
Hb_188303_010 |
0.3261292999 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 20 |
Hb_001310_050 |
0.3261529852 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |