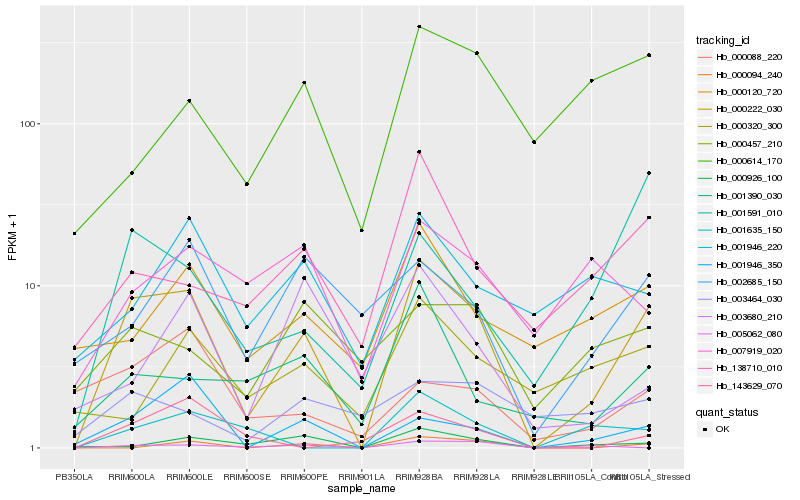
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000222_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_001946_350 |
0.2927901695 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |
| 3 |
Hb_000926_100 |
0.2990244085 |
- |
- |
hypothetical protein B456_007G076800 [Gossypium raimondii] |
| 4 |
Hb_138710_010 |
0.3115921839 |
- |
- |
Adenosylmethionine-8-amino-7-oxononanoate aminotransferase [Gossypium arboreum] |
| 5 |
Hb_000120_720 |
0.3150197208 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 6 |
Hb_002685_150 |
0.3174995783 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 7 |
Hb_001390_030 |
0.3182828667 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 8 |
Hb_001635_150 |
0.3259741103 |
- |
- |
hypothetical protein JCGZ_24809 [Jatropha curcas] |
| 9 |
Hb_001946_220 |
0.3261204568 |
- |
- |
PREDICTED: signal peptidase complex catalytic subunit SEC11A-like [Jatropha curcas] |
| 10 |
Hb_000094_240 |
0.3267155111 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 11 |
Hb_001591_010 |
0.3308587883 |
transcription factor |
TF Family: MYB |
myb domain protein [Hevea brasiliensis] |
| 12 |
Hb_000457_210 |
0.3353495377 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 13 |
Hb_000320_300 |
0.336173333 |
- |
- |
prephenate dehydratase, putative [Ricinus communis] |
| 14 |
Hb_000088_220 |
0.3388025871 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000614_170 |
0.3397490465 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 16 |
Hb_003464_030 |
0.3403451362 |
- |
- |
PREDICTED: 60S ribosomal protein L24-like [Jatropha curcas] |
| 17 |
Hb_005062_080 |
0.3444016071 |
- |
- |
MLO-like protein 4 [Gossypium arboreum] |
| 18 |
Hb_007919_020 |
0.3450314856 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 19 |
Hb_143629_070 |
0.3451619605 |
- |
- |
PREDICTED: uncharacterized protein LOC105647357 [Jatropha curcas] |
| 20 |
Hb_003680_210 |
0.3478698021 |
transcription factor |
TF Family: Trihelix |
PREDICTED: stress response protein NST1 [Jatropha curcas] |