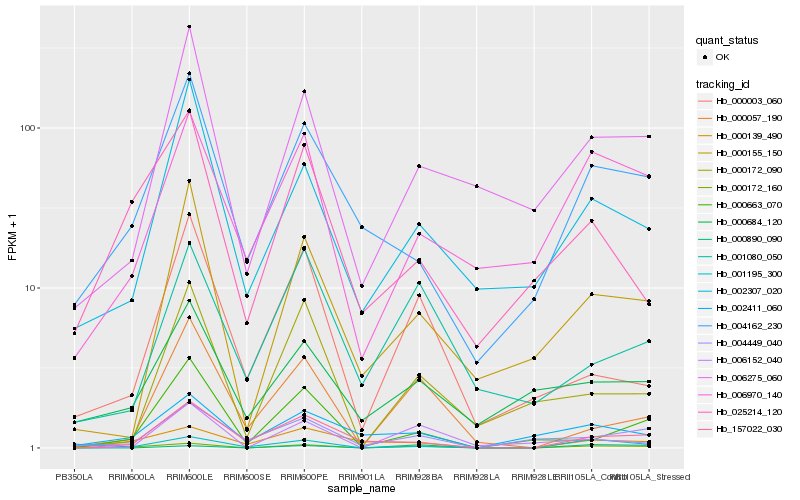
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000172_160 |
0.0 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 2 |
Hb_006970_140 |
0.2730645216 |
- |
- |
PREDICTED: histone H3.2-like [Solanum lycopersicum] |
| 3 |
Hb_000139_490 |
0.275681097 |
- |
- |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 4 |
Hb_006152_040 |
0.2843503302 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105645062 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000890_090 |
0.2848930702 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF086 [Jatropha curcas] |
| 6 |
Hb_000057_190 |
0.2856403755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_157022_030 |
0.2883323628 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 8 |
Hb_004449_040 |
0.2924423441 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 9 |
Hb_004162_230 |
0.2932103612 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 10 |
Hb_006275_060 |
0.2941995977 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 11 |
Hb_000663_070 |
0.2959780059 |
- |
- |
cyclin A, putative [Ricinus communis] |
| 12 |
Hb_000155_150 |
0.2963037286 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 13 |
Hb_002411_060 |
0.298455625 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 5 [Jatropha curcas] |
| 14 |
Hb_000172_090 |
0.3030879425 |
- |
- |
PREDICTED: uncharacterized protein LOC105650755 [Jatropha curcas] |
| 15 |
Hb_001080_050 |
0.3055521005 |
- |
- |
PREDICTED: uncharacterized protein LOC105635235 [Jatropha curcas] |
| 16 |
Hb_000684_120 |
0.3078787572 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002307_020 |
0.3081916783 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 18 |
Hb_001195_300 |
0.3091383885 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 19 |
Hb_025214_120 |
0.3130790364 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13 [Jatropha curcas] |
| 20 |
Hb_000003_060 |
0.3143229871 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |