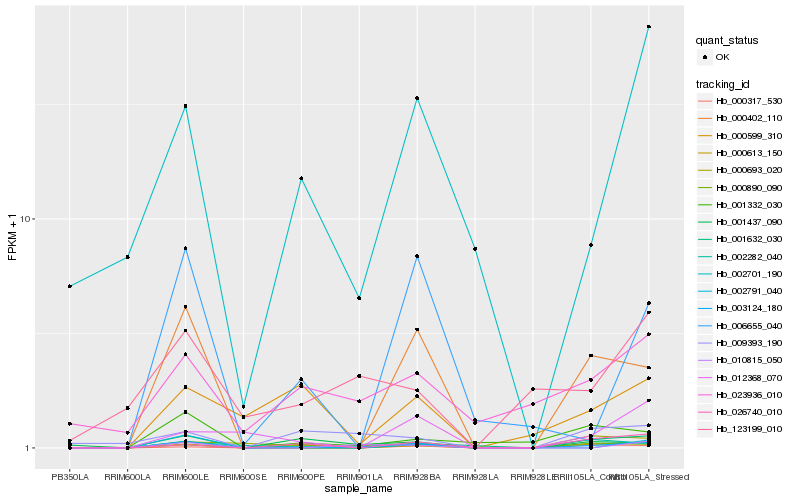
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003124_180 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000402_110 |
0.2582815239 |
- |
- |
hypothetical protein JCGZ_17786 [Jatropha curcas] |
| 3 |
Hb_000613_150 |
0.2654458821 |
- |
- |
PREDICTED: uncharacterized protein LOC105641537 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001632_030 |
0.2748074216 |
- |
- |
PREDICTED: chaperone protein ClpB1 [Jatropha curcas] |
| 5 |
Hb_002701_190 |
0.3067664854 |
- |
- |
PREDICTED: uncharacterized protein LOC105646742 [Jatropha curcas] |
| 6 |
Hb_000890_090 |
0.3091612246 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF086 [Jatropha curcas] |
| 7 |
Hb_006655_040 |
0.3234774366 |
- |
- |
tau class glutathione transferase GSTU15 [Populus trichocarpa] |
| 8 |
Hb_001332_030 |
0.3274499658 |
- |
- |
hypothetical protein JCGZ_21111 [Jatropha curcas] |
| 9 |
Hb_026740_010 |
0.3342581785 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 10 |
Hb_001437_090 |
0.3360607281 |
- |
- |
PREDICTED: phosphatidylinositol N-acetylglucosaminyltransferase gpi3 subunit [Jatropha curcas] |
| 11 |
Hb_023936_010 |
0.340467612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010815_050 |
0.3441482705 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 13 |
Hb_000317_530 |
0.3449341248 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 14 |
Hb_012368_070 |
0.3496578794 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_000693_020 |
0.3550812136 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 16 |
Hb_000599_310 |
0.3551742028 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8-like [Jatropha curcas] |
| 17 |
Hb_002282_040 |
0.3585699827 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 18 |
Hb_123199_010 |
0.3591606489 |
- |
- |
SRK protein [Brassica cretica] |
| 19 |
Hb_009393_190 |
0.3608493908 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |
| 20 |
Hb_002791_040 |
0.3645223693 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |