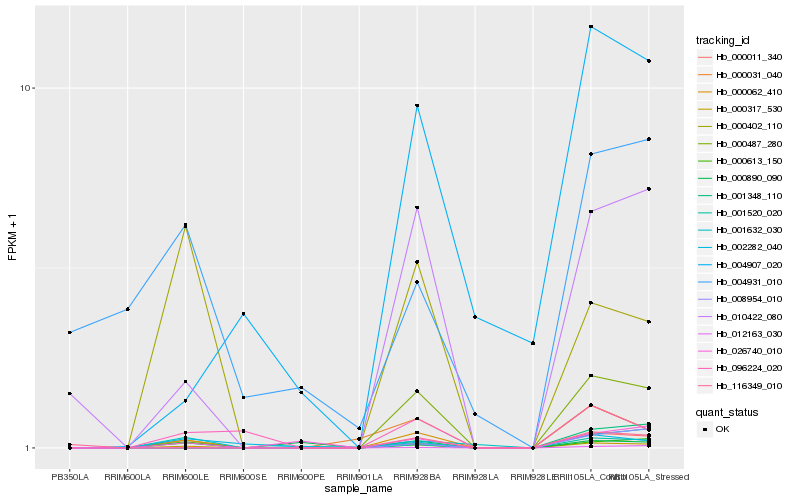
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000317_530 |
0.0 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 2 |
Hb_000062_410 |
0.1438386392 |
- |
- |
PREDICTED: uncharacterized protein LOC105644766 [Jatropha curcas] |
| 3 |
Hb_000613_150 |
0.182225869 |
- |
- |
PREDICTED: uncharacterized protein LOC105641537 isoform X1 [Jatropha curcas] |
| 4 |
Hb_010422_080 |
0.2007792917 |
- |
- |
- |
| 5 |
Hb_012163_030 |
0.2038645322 |
- |
- |
PREDICTED: uncharacterized protein LOC105647861 [Jatropha curcas] |
| 6 |
Hb_000031_040 |
0.2466122024 |
- |
- |
centromere protein O [Medicago truncatula] |
| 7 |
Hb_000487_280 |
0.2576123447 |
- |
- |
- |
| 8 |
Hb_001632_030 |
0.2777509742 |
- |
- |
PREDICTED: chaperone protein ClpB1 [Jatropha curcas] |
| 9 |
Hb_000011_340 |
0.2782926011 |
- |
- |
PREDICTED: protein LURP-one-related 11-like [Jatropha curcas] |
| 10 |
Hb_026740_010 |
0.2786854528 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 11 |
Hb_001520_020 |
0.2790445361 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 12 |
Hb_096224_020 |
0.2904399251 |
- |
- |
hypothetical protein JCGZ_06980 [Jatropha curcas] |
| 13 |
Hb_000402_110 |
0.2944641273 |
- |
- |
hypothetical protein JCGZ_17786 [Jatropha curcas] |
| 14 |
Hb_002282_040 |
0.3079491366 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 15 |
Hb_004907_020 |
0.3120662199 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |
| 16 |
Hb_116349_010 |
0.3191395569 |
- |
- |
PREDICTED: uncharacterized protein LOC105763153 [Gossypium raimondii] |
| 17 |
Hb_008954_010 |
0.3271658105 |
- |
- |
- |
| 18 |
Hb_000890_090 |
0.3279985433 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF086 [Jatropha curcas] |
| 19 |
Hb_001348_110 |
0.3364346551 |
- |
- |
PREDICTED: esterase-like [Jatropha curcas] |
| 20 |
Hb_004931_010 |
0.3447115806 |
- |
- |
inosine triphosphate pyrophosphatase, putative [Ricinus communis] |