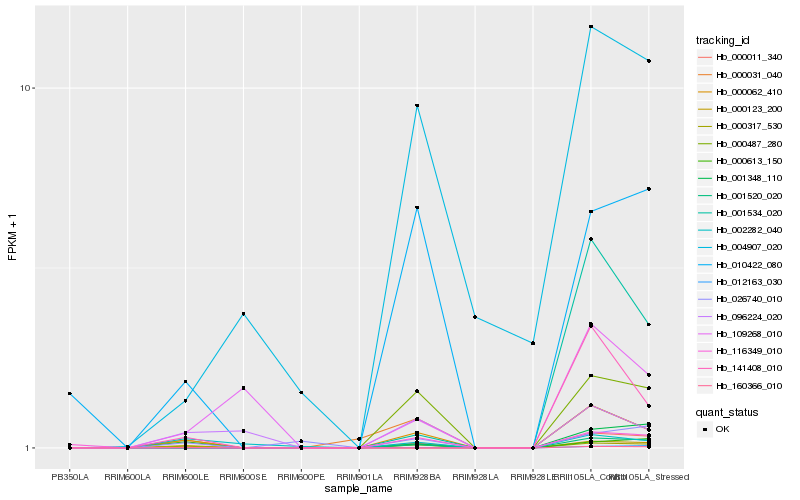
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_410 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644766 [Jatropha curcas] |
| 2 |
Hb_000317_530 |
0.1438386392 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 3 |
Hb_000031_040 |
0.2357879413 |
- |
- |
centromere protein O [Medicago truncatula] |
| 4 |
Hb_000011_340 |
0.249717566 |
- |
- |
PREDICTED: protein LURP-one-related 11-like [Jatropha curcas] |
| 5 |
Hb_001520_020 |
0.2501265575 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 6 |
Hb_000487_280 |
0.2633324234 |
- |
- |
- |
| 7 |
Hb_012163_030 |
0.2674066453 |
- |
- |
PREDICTED: uncharacterized protein LOC105647861 [Jatropha curcas] |
| 8 |
Hb_010422_080 |
0.2691339196 |
- |
- |
- |
| 9 |
Hb_000613_150 |
0.2935826098 |
- |
- |
PREDICTED: uncharacterized protein LOC105641537 isoform X1 [Jatropha curcas] |
| 10 |
Hb_096224_020 |
0.2966193712 |
- |
- |
hypothetical protein JCGZ_06980 [Jatropha curcas] |
| 11 |
Hb_004907_020 |
0.3120798396 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |
| 12 |
Hb_116349_010 |
0.3182021116 |
- |
- |
PREDICTED: uncharacterized protein LOC105763153 [Gossypium raimondii] |
| 13 |
Hb_001534_020 |
0.3242873655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_141408_010 |
0.3270455494 |
- |
- |
PREDICTED: uncharacterized protein LOC105631382 [Jatropha curcas] |
| 15 |
Hb_002282_040 |
0.3289222442 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 16 |
Hb_001348_110 |
0.3350969646 |
- |
- |
PREDICTED: esterase-like [Jatropha curcas] |
| 17 |
Hb_160366_010 |
0.3352033822 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_000123_200 |
0.3366254961 |
- |
- |
PREDICTED: WAT1-related protein At2g39510 [Jatropha curcas] |
| 19 |
Hb_109268_010 |
0.3368629477 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 20 |
Hb_026740_010 |
0.3396207192 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |