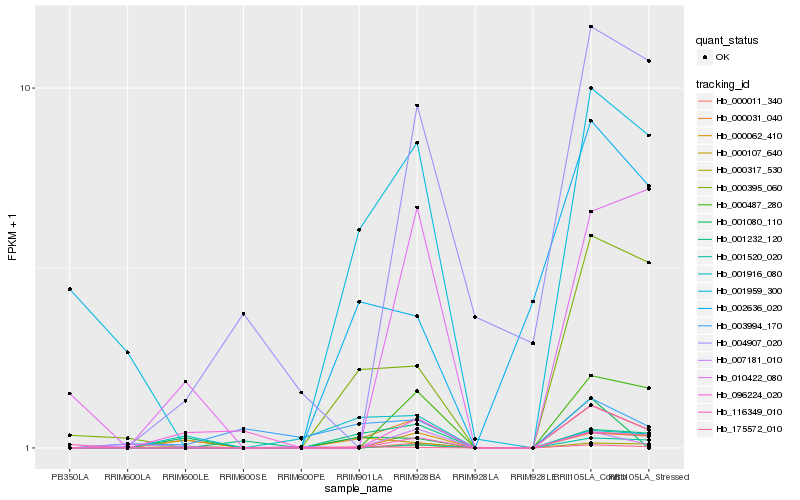
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000031_040 |
0.0 |
- |
- |
centromere protein O [Medicago truncatula] |
| 2 |
Hb_000062_410 |
0.2357879413 |
- |
- |
PREDICTED: uncharacterized protein LOC105644766 [Jatropha curcas] |
| 3 |
Hb_000317_530 |
0.2466122024 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 4 |
Hb_010422_080 |
0.302037415 |
- |
- |
- |
| 5 |
Hb_000487_280 |
0.3027716815 |
- |
- |
- |
| 6 |
Hb_001959_300 |
0.3085325191 |
- |
- |
PREDICTED: 3-isopropylmalate dehydratase large subunit [Jatropha curcas] |
| 7 |
Hb_000395_060 |
0.3133544624 |
- |
- |
- |
| 8 |
Hb_000107_640 |
0.317144226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_175572_010 |
0.317464846 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 10 |
Hb_001232_120 |
0.3329884853 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16010 [Jatropha curcas] |
| 11 |
Hb_096224_020 |
0.3380548153 |
- |
- |
hypothetical protein JCGZ_06980 [Jatropha curcas] |
| 12 |
Hb_000011_340 |
0.3440731183 |
- |
- |
PREDICTED: protein LURP-one-related 11-like [Jatropha curcas] |
| 13 |
Hb_001520_020 |
0.3450085253 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 14 |
Hb_007181_010 |
0.3571600103 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 15 |
Hb_116349_010 |
0.3591498995 |
- |
- |
PREDICTED: uncharacterized protein LOC105763153 [Gossypium raimondii] |
| 16 |
Hb_004907_020 |
0.360319738 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |
| 17 |
Hb_001916_080 |
0.3606012493 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 18 |
Hb_003994_170 |
0.3639186547 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 19 |
Hb_002636_020 |
0.3647394353 |
- |
- |
- |
| 20 |
Hb_001080_110 |
0.3674483766 |
- |
- |
- |