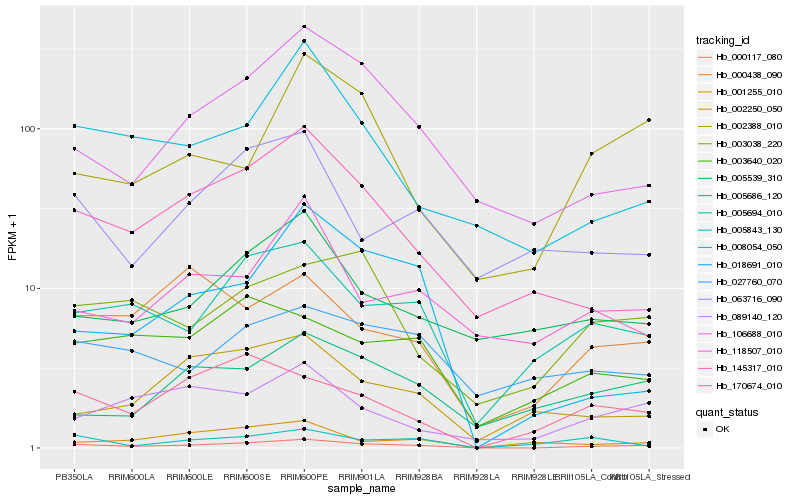
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000117_080 |
0.0 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 2 |
Hb_005694_010 |
0.1876757657 |
transcription factor |
TF Family: C2C2-Dof |
F-box family protein, putative isoform 1 [Theobroma cacao] |
| 3 |
Hb_106688_010 |
0.2102958515 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 4 |
Hb_170674_010 |
0.214735948 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 5 |
Hb_003038_220 |
0.220220529 |
- |
- |
eukaryotic translation initiation factor 2 gamma subunit, putative [Ricinus communis] |
| 6 |
Hb_008054_050 |
0.2214929949 |
- |
- |
PREDICTED: uncharacterized protein LOC101212042 [Cucumis sativus] |
| 7 |
Hb_003640_020 |
0.223002509 |
- |
- |
pelota, putative [Ricinus communis] |
| 8 |
Hb_089140_120 |
0.2232769053 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 9 |
Hb_000438_090 |
0.2266822168 |
- |
- |
PREDICTED: mini-chromosome maintenance complex-binding protein [Jatropha curcas] |
| 10 |
Hb_145317_010 |
0.2276871278 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 11 [Jatropha curcas] |
| 11 |
Hb_002388_010 |
0.2282120692 |
- |
- |
- |
| 12 |
Hb_005686_120 |
0.2333247976 |
- |
- |
- |
| 13 |
Hb_005539_310 |
0.2347706379 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 14 |
Hb_001255_010 |
0.2355522728 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Tarenaya hassleriana] |
| 15 |
Hb_063716_090 |
0.2359131161 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_018691_010 |
0.2364446709 |
- |
- |
- |
| 17 |
Hb_002250_050 |
0.2372121905 |
- |
- |
PREDICTED: uncharacterized protein LOC105638972 [Jatropha curcas] |
| 18 |
Hb_027760_070 |
0.2381994272 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 19 |
Hb_118507_010 |
0.2383815821 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 20 |
Hb_005843_130 |
0.2385316651 |
- |
- |
hypothetical protein JCGZ_12389 [Jatropha curcas] |