| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
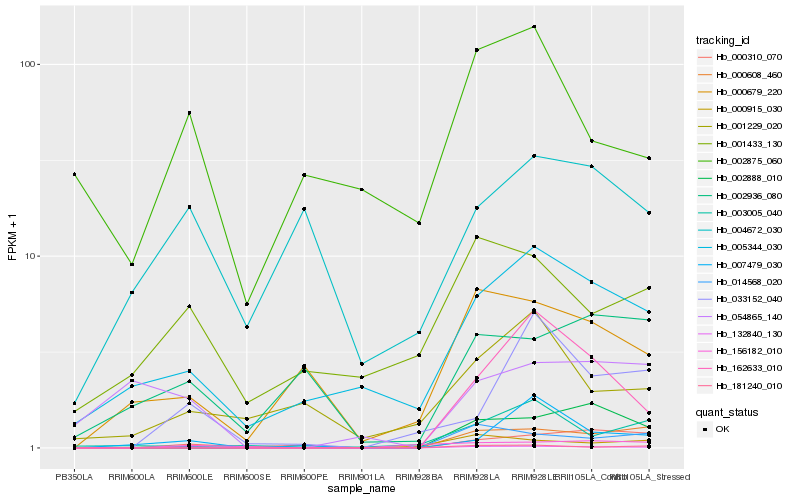
Hb_132840_130 |
0.0 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 8 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_000310_070 |
0.3045867948 |
- |
- |
BnaA06g08330D [Brassica napus] |
| 3 |
Hb_005344_030 |
0.3142180825 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Jatropha curcas] |
| 4 |
Hb_033152_040 |
0.3211556587 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 5 |
Hb_001433_130 |
0.3245038179 |
- |
- |
PREDICTED: uncharacterized protein LOC105647468 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000679_220 |
0.3246924356 |
- |
- |
PREDICTED: uncharacterized protein LOC105637638 [Jatropha curcas] |
| 7 |
Hb_156182_010 |
0.3259404547 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 8 |
Hb_000608_460 |
0.327126175 |
- |
- |
- |
| 9 |
Hb_014568_020 |
0.3277212269 |
- |
- |
PREDICTED: GPI mannosyltransferase 2 [Jatropha curcas] |
| 10 |
Hb_002888_010 |
0.3425475123 |
- |
- |
- |
| 11 |
Hb_007479_030 |
0.350106396 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 12 |
Hb_002936_080 |
0.351195206 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 13 |
Hb_002875_060 |
0.3537746664 |
- |
- |
hypothetical protein L484_023461 [Morus notabilis] |
| 14 |
Hb_054865_140 |
0.3543665533 |
- |
- |
- |
| 15 |
Hb_000915_030 |
0.3546982874 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 16 |
Hb_162633_010 |
0.3599904276 |
- |
- |
hypothetical protein POPTR_0011s02600g [Populus trichocarpa] |
| 17 |
Hb_001229_020 |
0.3635468066 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 18 |
Hb_003005_040 |
0.3638670071 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_004672_030 |
0.3646509523 |
- |
- |
proline oxidase [Manihot esculenta] |
| 20 |
Hb_181240_010 |
0.3654072031 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |