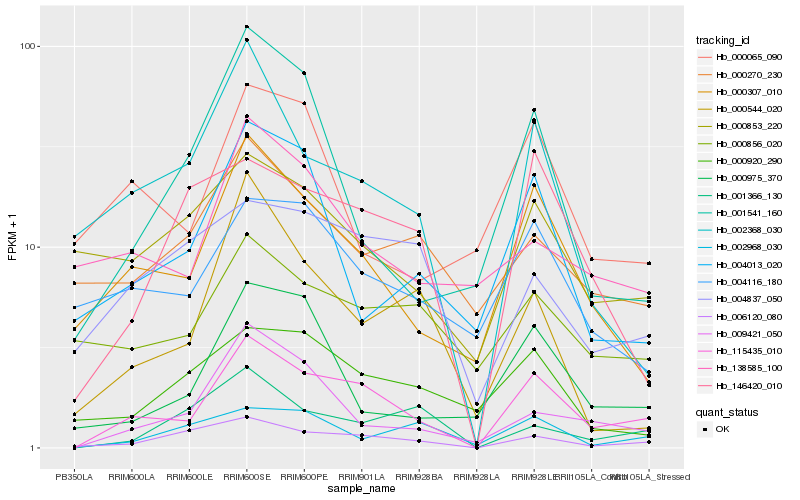
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_115435_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000307_010 |
0.1999832026 |
- |
- |
PREDICTED: ganglioside-induced differentiation-associated protein 2-like [Jatropha curcas] |
| 3 |
Hb_006120_080 |
0.2054773629 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48810 [Jatropha curcas] |
| 4 |
Hb_009421_050 |
0.2347591366 |
- |
- |
PREDICTED: uncharacterized protein LOC105647037 [Jatropha curcas] |
| 5 |
Hb_002368_030 |
0.2400478336 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 6 |
Hb_146420_010 |
0.2415543 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_000975_370 |
0.2503027819 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB59 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004116_180 |
0.2523741425 |
- |
- |
PREDICTED: uncharacterized protein LOC105646147 [Jatropha curcas] |
| 9 |
Hb_004013_020 |
0.2546339984 |
- |
- |
PREDICTED: F-box protein SKIP2 [Jatropha curcas] |
| 10 |
Hb_000856_020 |
0.2561003547 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 11 |
Hb_000853_220 |
0.2570071306 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 12 |
Hb_004837_050 |
0.2578750911 |
- |
- |
- |
| 13 |
Hb_001366_130 |
0.2579891819 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_000544_020 |
0.2592246649 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 15 |
Hb_000920_290 |
0.2600286514 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 16 |
Hb_001541_160 |
0.2609320464 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 17 |
Hb_138585_100 |
0.2626407775 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 18 |
Hb_000270_230 |
0.2627498627 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 19 |
Hb_002968_030 |
0.2638986298 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000065_090 |
0.2648470238 |
- |
- |
potassium channel tetramerization domain-containing protein, putative [Ricinus communis] |