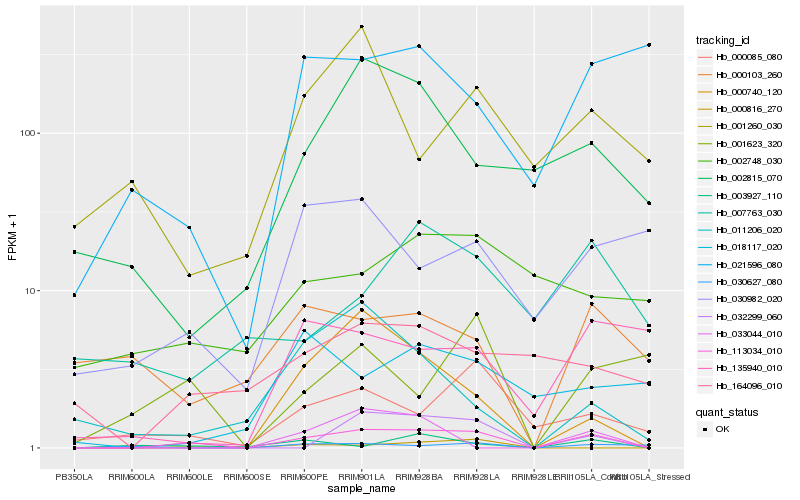
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_113034_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_032299_060 |
0.3309616472 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC-like [Elaeis guineensis] |
| 3 |
Hb_000740_120 |
0.3360056647 |
- |
- |
- |
| 4 |
Hb_000085_080 |
0.3467616147 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 1 [Jatropha curcas] |
| 5 |
Hb_000816_270 |
0.3516061397 |
- |
- |
- |
| 6 |
Hb_003927_110 |
0.3682179631 |
- |
- |
hypothetical protein PRUPE_ppa001478mg [Prunus persica] |
| 7 |
Hb_002815_070 |
0.3693365002 |
- |
- |
hypothetical protein PHAVU_005G048300g [Phaseolus vulgaris] |
| 8 |
Hb_011206_020 |
0.3796803123 |
- |
- |
- |
| 9 |
Hb_030627_080 |
0.3814085155 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_164096_010 |
0.3831307406 |
- |
- |
hypothetical protein L484_021244 [Morus notabilis] |
| 11 |
Hb_001623_320 |
0.3908707122 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 isoform X1 [Populus euphratica] |
| 12 |
Hb_033044_010 |
0.3926329484 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 5-like isoform X3 [Gossypium raimondii] |
| 13 |
Hb_000103_260 |
0.3948719193 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 14 |
Hb_135940_010 |
0.3955805317 |
- |
- |
hypothetical protein JCGZ_18294 [Jatropha curcas] |
| 15 |
Hb_030982_020 |
0.3960099792 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1 [Jatropha curcas] |
| 16 |
Hb_007763_030 |
0.3969033007 |
- |
- |
PREDICTED: uncharacterized protein LOC105638154 [Jatropha curcas] |
| 17 |
Hb_001260_030 |
0.3989757585 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002748_030 |
0.4003265112 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 19 |
Hb_018117_020 |
0.403817309 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 20 |
Hb_021596_080 |
0.4044970492 |
- |
- |
50S ribosomal protein L33A [Hevea brasiliensis] |