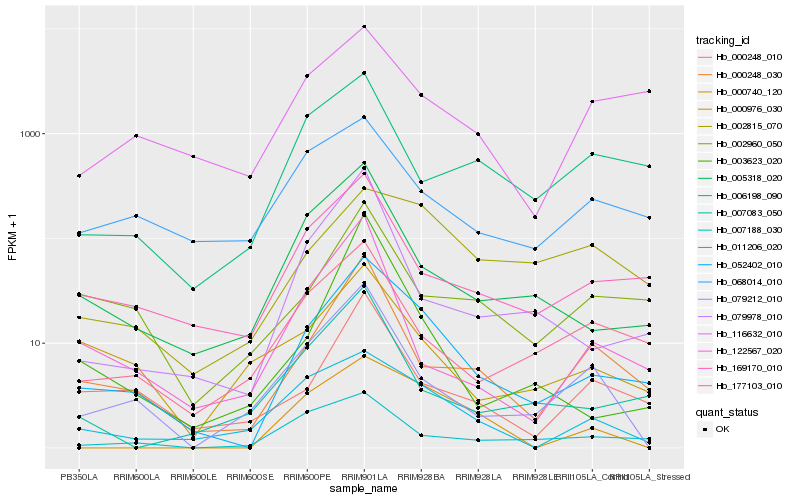
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052402_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_169170_010 |
0.2008367194 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 3 |
Hb_005318_020 |
0.2049953516 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000248_030 |
0.2078904401 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 5 |
Hb_011206_020 |
0.2201662431 |
- |
- |
- |
| 6 |
Hb_177103_010 |
0.2246341277 |
- |
- |
PREDICTED: histone H2AX [Elaeis guineensis] |
| 7 |
Hb_000248_010 |
0.2258008455 |
- |
- |
hypothetical protein PRUPE_ppa008056mg [Prunus persica] |
| 8 |
Hb_079978_010 |
0.2316916693 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP71 isoform X1 [Nelumbo nucifera] |
| 9 |
Hb_002960_050 |
0.2320853587 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 10 |
Hb_007188_030 |
0.2344017572 |
- |
- |
PREDICTED: DNA topoisomerase 3-alpha-like [Malus domestica] |
| 11 |
Hb_007083_050 |
0.236871897 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X2 [Nelumbo nucifera] |
| 12 |
Hb_000976_030 |
0.2373493803 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase-like [Citrus sinensis] |
| 13 |
Hb_116632_010 |
0.2432723888 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_079212_010 |
0.2445438717 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_068014_010 |
0.2457293578 |
- |
- |
hypothetical protein POPTR_0312s00200g, partial [Populus trichocarpa] |
| 16 |
Hb_000740_120 |
0.2484735098 |
- |
- |
- |
| 17 |
Hb_002815_070 |
0.2490233646 |
- |
- |
hypothetical protein PHAVU_005G048300g [Phaseolus vulgaris] |
| 18 |
Hb_003623_020 |
0.252598325 |
- |
- |
- |
| 19 |
Hb_006198_090 |
0.254757184 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |
| 20 |
Hb_122567_020 |
0.2564951132 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |