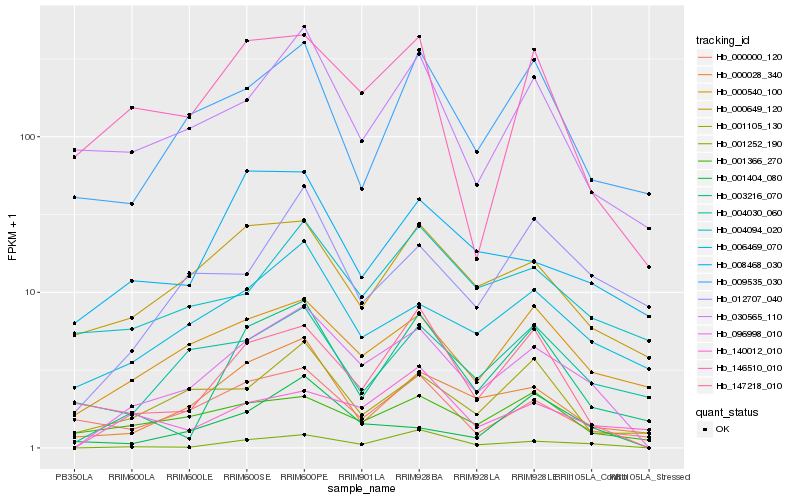
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_096998_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_07944 [Jatropha curcas] |
| 2 |
Hb_001252_190 |
0.1752125451 |
desease resistance |
Gene Name: NB-ARC |
- |
| 3 |
Hb_000540_100 |
0.1946119186 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g60050 [Jatropha curcas] |
| 4 |
Hb_006469_070 |
0.1959703664 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040 [Jatropha curcas] |
| 5 |
Hb_000028_340 |
0.1962925612 |
- |
- |
PREDICTED: potassium channel SKOR-like [Prunus mume] |
| 6 |
Hb_000649_120 |
0.2059966895 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_003216_070 |
0.2070844456 |
- |
- |
hypothetical protein RCOM_0483300 [Ricinus communis] |
| 8 |
Hb_030565_110 |
0.214573468 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 9 |
Hb_140012_010 |
0.2169469091 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Eucalyptus grandis] |
| 10 |
Hb_001105_130 |
0.2171829957 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 11 |
Hb_147218_010 |
0.2192205928 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 12 |
Hb_001366_270 |
0.2196237215 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 13 |
Hb_009535_030 |
0.2208970435 |
- |
- |
CP2 [Hevea brasiliensis] |
| 14 |
Hb_012707_040 |
0.2209357844 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 29 [Jatropha curcas] |
| 15 |
Hb_001404_080 |
0.221746528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000000_120 |
0.2219479981 |
- |
- |
hypothetical protein EUGRSUZ_A004731, partial [Eucalyptus grandis] |
| 17 |
Hb_146510_010 |
0.2244594114 |
- |
- |
cinnamyl alcohol dehydrogenase [Hevea brasiliensis] |
| 18 |
Hb_004094_020 |
0.2247798115 |
- |
- |
Pyruvate kinase family protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_008468_030 |
0.2259548338 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 20 |
Hb_004030_060 |
0.2264383817 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |