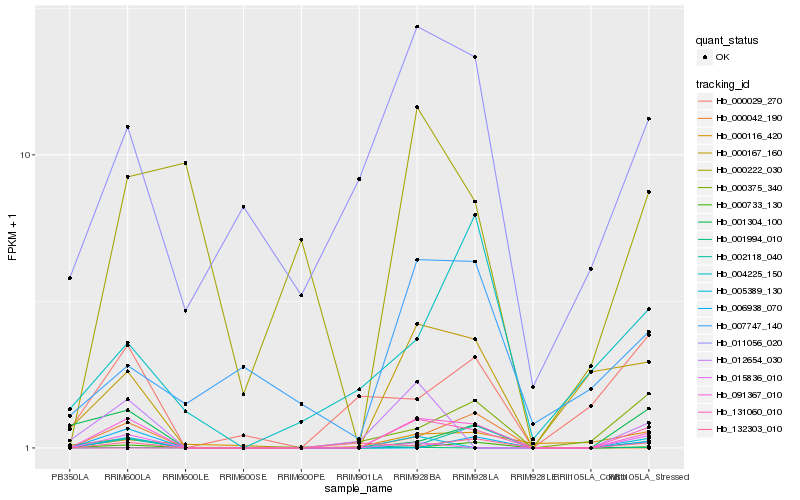
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_091367_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_18029 [Jatropha curcas] |
| 2 |
Hb_000167_160 |
0.3279874837 |
- |
- |
- |
| 3 |
Hb_002118_040 |
0.3410875487 |
- |
- |
PREDICTED: uncharacterized protein LOC104727990 [Camelina sativa] |
| 4 |
Hb_000042_190 |
0.3585404959 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005389_130 |
0.3782950941 |
- |
- |
hypothetical protein CICLE_v10009852mg [Citrus clementina] |
| 6 |
Hb_011056_020 |
0.3953221229 |
transcription factor |
TF Family: ERF |
AP2 domain transcription factor RAP2.3, putative [Ricinus communis] |
| 7 |
Hb_132303_010 |
0.4011964566 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 8 |
Hb_012654_030 |
0.4129371506 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000116_420 |
0.4154002744 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2-like [Jatropha curcas] |
| 10 |
Hb_000029_270 |
0.4162253167 |
- |
- |
PREDICTED: uncharacterized protein LOC105634361 [Jatropha curcas] |
| 11 |
Hb_007747_140 |
0.4183739715 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_001994_010 |
0.4192337779 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_006938_070 |
0.4225013708 |
- |
- |
PREDICTED: uncharacterized protein LOC104893877 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_000222_030 |
0.4230053072 |
- |
- |
- |
| 15 |
Hb_131060_010 |
0.4257668942 |
- |
- |
PREDICTED: uncharacterized protein LOC105800955 [Gossypium raimondii] |
| 16 |
Hb_000733_130 |
0.4332684804 |
- |
- |
PREDICTED: cucumisin [Vitis vinifera] |
| 17 |
Hb_004225_150 |
0.4342265379 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger protein 36, C3H1 type-like 3 [Jatropha curcas] |
| 18 |
Hb_015836_010 |
0.4343588152 |
- |
- |
- |
| 19 |
Hb_000375_340 |
0.4352352119 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Populus euphratica] |
| 20 |
Hb_001304_100 |
0.4364893776 |
- |
- |
- |