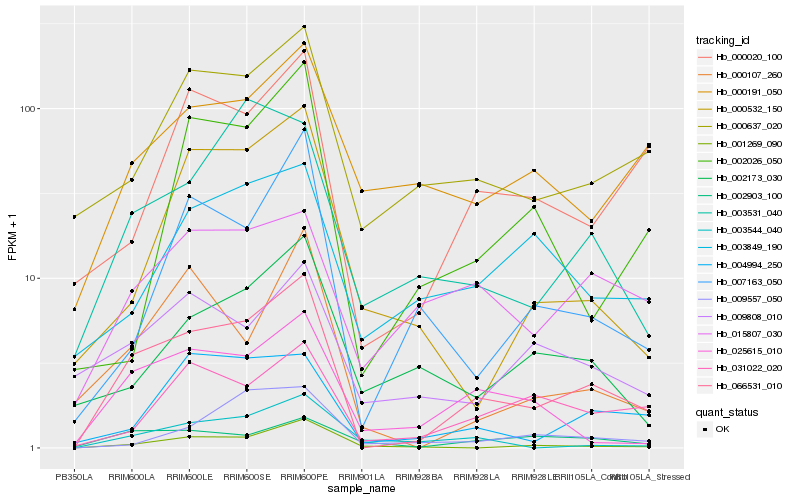
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_066531_010 |
0.0 |
- |
- |
ccd1, putative [Ricinus communis] |
| 2 |
Hb_025615_010 |
0.2256275204 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 3 |
Hb_000637_020 |
0.2304640112 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 4 |
Hb_000020_100 |
0.230933443 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 5 |
Hb_001269_090 |
0.2316626229 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 6 |
Hb_004994_250 |
0.2368707349 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 7 |
Hb_000107_260 |
0.2418853875 |
- |
- |
- |
| 8 |
Hb_007163_050 |
0.2431404047 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9 [Jatropha curcas] |
| 9 |
Hb_003531_040 |
0.2450363026 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 10 |
Hb_002903_100 |
0.2474941523 |
- |
- |
hypothetical protein POPTR_0011s04750g [Populus trichocarpa] |
| 11 |
Hb_031022_020 |
0.249870404 |
- |
- |
- |
| 12 |
Hb_003544_040 |
0.2504791619 |
- |
- |
PREDICTED: protein TOO MANY MOUTHS [Jatropha curcas] |
| 13 |
Hb_015807_030 |
0.2536447609 |
- |
- |
PREDICTED: protein NDR1-like [Jatropha curcas] |
| 14 |
Hb_000532_150 |
0.2539794368 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 15 |
Hb_009808_010 |
0.2573202098 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_002173_030 |
0.2574289107 |
- |
- |
hypothetical protein JCGZ_12466 [Jatropha curcas] |
| 17 |
Hb_000191_050 |
0.2579693082 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |
| 18 |
Hb_002026_050 |
0.2622476072 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 19 |
Hb_009557_050 |
0.2640896097 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 20 |
Hb_003849_190 |
0.2656682801 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |