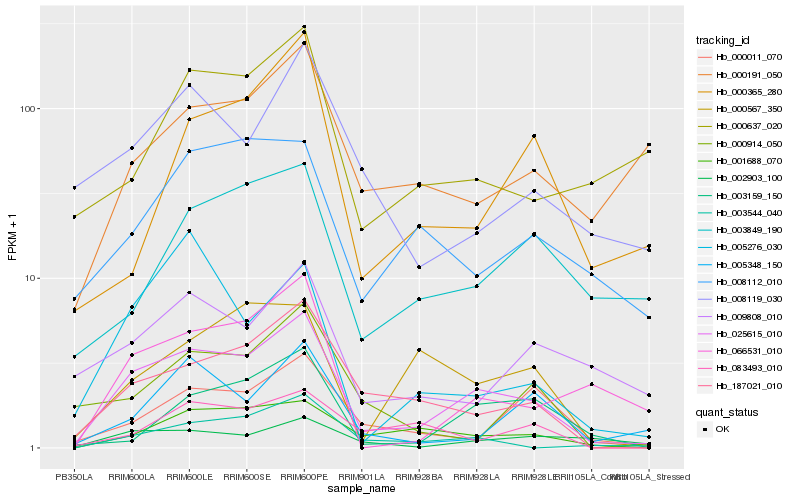
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025615_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 2 |
Hb_187021_010 |
0.1914291502 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 3 |
Hb_001688_070 |
0.2125406047 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X1 [Jatropha curcas] |
| 4 |
Hb_003544_040 |
0.2201156366 |
- |
- |
PREDICTED: protein TOO MANY MOUTHS [Jatropha curcas] |
| 5 |
Hb_066531_010 |
0.2256275204 |
- |
- |
ccd1, putative [Ricinus communis] |
| 6 |
Hb_000567_350 |
0.2390390031 |
- |
- |
PREDICTED: probable acyl-activating enzyme 6 [Jatropha curcas] |
| 7 |
Hb_008112_010 |
0.2396140963 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 8 |
Hb_000011_070 |
0.2412874587 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 9 |
Hb_008119_030 |
0.2424039306 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 10 |
Hb_002903_100 |
0.2432674829 |
- |
- |
hypothetical protein POPTR_0011s04750g [Populus trichocarpa] |
| 11 |
Hb_005348_150 |
0.2457597362 |
- |
- |
PREDICTED: actin-100-like [Jatropha curcas] |
| 12 |
Hb_003159_150 |
0.2465913934 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 8 [Jatropha curcas] |
| 13 |
Hb_083493_010 |
0.2482504361 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 14 |
Hb_009808_010 |
0.248413004 |
- |
- |
Leucine-rich repeat protein kinase family protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_000191_050 |
0.2488115158 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |
| 16 |
Hb_003849_190 |
0.2505242574 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 17 |
Hb_000637_020 |
0.2513159621 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 18 |
Hb_000365_280 |
0.2515531 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 19 |
Hb_000914_050 |
0.2531713962 |
- |
- |
PREDICTED: serine/threonine-protein kinase UCNL [Jatropha curcas] |
| 20 |
Hb_005276_030 |
0.2551030585 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |