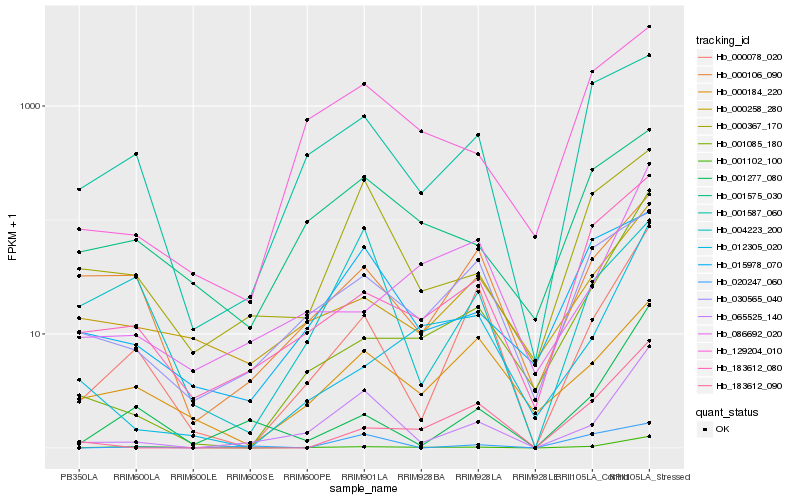
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065525_140 |
0.0 |
- |
- |
PREDICTED: 50S ribosomal protein L12, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000078_020 |
0.2261728997 |
- |
- |
- |
| 3 |
Hb_000367_170 |
0.2463743456 |
- |
- |
PREDICTED: LYR motif-containing protein 4 [Jatropha curcas] |
| 4 |
Hb_001102_100 |
0.257523679 |
- |
- |
PREDICTED: uncharacterized protein LOC105140429 isoform X2 [Populus euphratica] |
| 5 |
Hb_129204_010 |
0.2603749289 |
- |
- |
LOC100284182 [Zea mays] |
| 6 |
Hb_001277_080 |
0.2711911657 |
- |
- |
PREDICTED: vegetative cell wall protein gp1 [Jatropha curcas] |
| 7 |
Hb_001085_180 |
0.2739502593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_183612_080 |
0.2804573708 |
- |
- |
hypothetical protein POPTR_0001s00500g [Populus trichocarpa] |
| 9 |
Hb_015978_070 |
0.2834725284 |
- |
- |
- |
| 10 |
Hb_020247_060 |
0.2835206695 |
- |
- |
UDP-glucoronosyl/UDP-glucosyl transferase family protein [Populus trichocarpa] |
| 11 |
Hb_000258_280 |
0.286309616 |
- |
- |
PREDICTED: probable ran guanine nucleotide release factor [Jatropha curcas] |
| 12 |
Hb_030565_040 |
0.2910945634 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 13 |
Hb_001587_060 |
0.2949351762 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001575_030 |
0.2972459668 |
- |
- |
hypothetical protein POPTR_0022s00880g [Populus trichocarpa] |
| 15 |
Hb_004223_200 |
0.2991888697 |
- |
- |
PREDICTED: auxin-induced protein 6B-like [Cucumis sativus] |
| 16 |
Hb_086692_020 |
0.3014954534 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_000106_090 |
0.3034026146 |
- |
- |
PREDICTED: uncharacterized protein LOC105634795 [Jatropha curcas] |
| 18 |
Hb_000184_220 |
0.3036799068 |
- |
- |
hypothetical protein CISIN_1g0207781mg [Citrus sinensis] |
| 19 |
Hb_012305_020 |
0.3039223599 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |
| 20 |
Hb_183612_090 |
0.306143486 |
- |
- |
- |