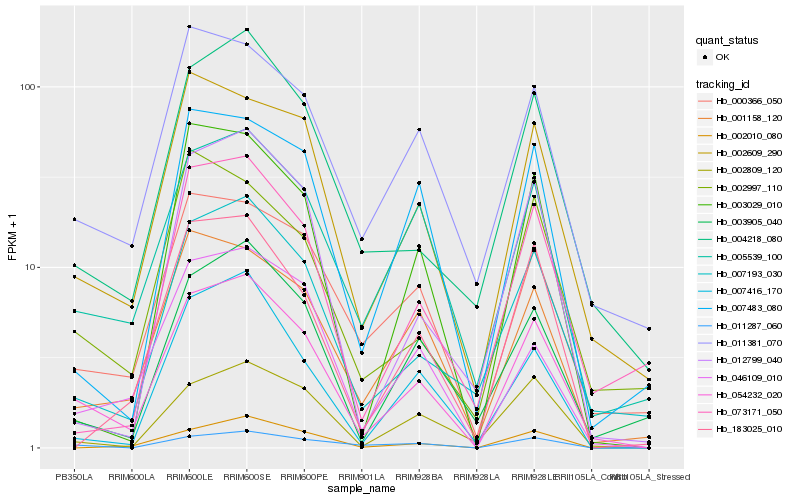
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054232_020 |
0.0 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 2 |
Hb_002609_290 |
0.1308521984 |
- |
- |
PREDICTED: non-specific phospholipase C1 [Jatropha curcas] |
| 3 |
Hb_007193_030 |
0.1326229169 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004218_080 |
0.1372595957 |
- |
- |
PREDICTED: uncharacterized protein LOC105631207 [Jatropha curcas] |
| 5 |
Hb_005539_100 |
0.1379339124 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 6 |
Hb_011381_070 |
0.1471428869 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 7 |
Hb_002809_120 |
0.1481535014 |
- |
- |
Uncharacterized protein TCM_031281 [Theobroma cacao] |
| 8 |
Hb_000366_050 |
0.1483233562 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 9 |
Hb_001158_120 |
0.1499828268 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003905_040 |
0.1532162835 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 11 |
Hb_011287_060 |
0.1548369887 |
- |
- |
- |
| 12 |
Hb_046109_010 |
0.1551778104 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 13 |
Hb_007416_170 |
0.1577971197 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 17 [Jatropha curcas] |
| 14 |
Hb_003029_010 |
0.1595463389 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_012799_040 |
0.161878278 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 16 |
Hb_183025_010 |
0.1626887326 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 17 |
Hb_002010_080 |
0.1628830754 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 18 |
Hb_002997_110 |
0.1641649562 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 22 isoform X1 [Jatropha curcas] |
| 19 |
Hb_073171_050 |
0.1651922973 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 20 |
Hb_007483_080 |
0.1652125577 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |