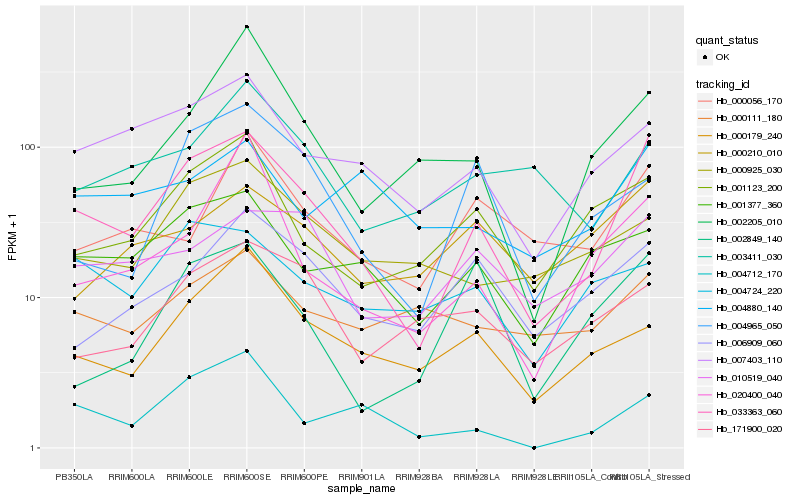
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033363_060 |
0.0 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 2 |
Hb_007403_110 |
0.1928125306 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |
| 3 |
Hb_001123_200 |
0.2032587004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001377_360 |
0.2033857686 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 5 |
Hb_010519_040 |
0.2131801488 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 6 |
Hb_004712_170 |
0.2190726853 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 7 |
Hb_000179_240 |
0.2199774384 |
- |
- |
PREDICTED: threonine synthase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_006909_060 |
0.2220389533 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3-like [Solanum lycopersicum] |
| 9 |
Hb_000925_030 |
0.2235524957 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 10 |
Hb_020400_040 |
0.2252289457 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181-like [Jatropha curcas] |
| 11 |
Hb_002205_010 |
0.225672432 |
- |
- |
hypothetical protein JCGZ_03337 [Jatropha curcas] |
| 12 |
Hb_000056_170 |
0.22594226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004724_220 |
0.228922645 |
- |
- |
PREDICTED: probable calcium-binding protein CML48 [Jatropha curcas] |
| 14 |
Hb_004965_050 |
0.230036848 |
- |
- |
PREDICTED: uncharacterized protein LOC105645055 [Jatropha curcas] |
| 15 |
Hb_000210_010 |
0.231439561 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 16 |
Hb_003411_030 |
0.2339555309 |
- |
- |
hypothetical protein POPTR_0010s13590g [Populus trichocarpa] |
| 17 |
Hb_002849_140 |
0.2342265656 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 18 |
Hb_171900_020 |
0.2344094414 |
- |
- |
PREDICTED: uncharacterized protein LOC105643207 [Jatropha curcas] |
| 19 |
Hb_000111_180 |
0.2347722036 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 20 |
Hb_004880_140 |
0.2351215581 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |