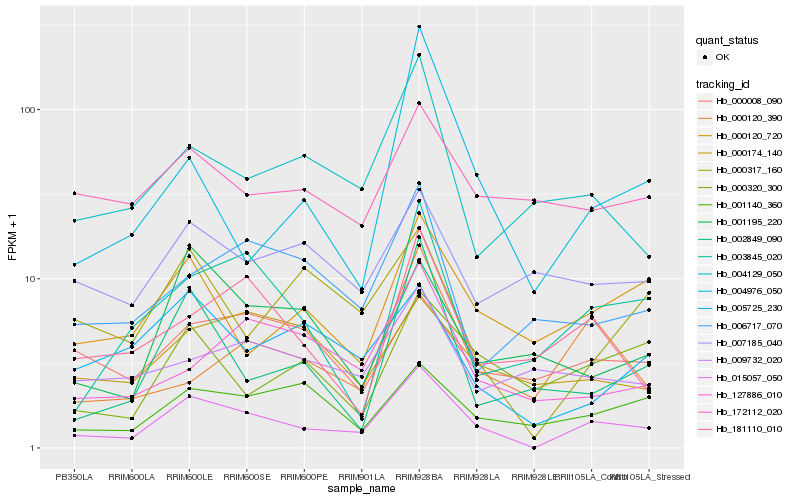
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015057_050 |
0.0 |
- |
- |
D-alanine aminotransferase, putative [Ricinus communis] |
| 2 |
Hb_000120_720 |
0.229483198 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 3 |
Hb_003845_020 |
0.2329641375 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_004129_050 |
0.2371123862 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 5 [Jatropha curcas] |
| 5 |
Hb_181110_010 |
0.237791933 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 6 |
Hb_000317_160 |
0.2404123876 |
- |
- |
2-nonaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [Theobroma cacao] |
| 7 |
Hb_127886_010 |
0.2415623109 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 8 |
Hb_000320_300 |
0.2422204541 |
- |
- |
prephenate dehydratase, putative [Ricinus communis] |
| 9 |
Hb_001140_360 |
0.2426827893 |
- |
- |
- |
| 10 |
Hb_005725_230 |
0.2454931724 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 11 |
Hb_172112_020 |
0.2467011095 |
- |
- |
PREDICTED: fumarylacetoacetase [Jatropha curcas] |
| 12 |
Hb_000120_390 |
0.246960345 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 13 |
Hb_006717_070 |
0.2470562245 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 14 |
Hb_002849_090 |
0.2491622476 |
- |
- |
hypothetical protein POPTR_0002s16380g [Populus trichocarpa] |
| 15 |
Hb_001195_220 |
0.2504996589 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 16 |
Hb_000008_090 |
0.2517285016 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 17 |
Hb_009732_020 |
0.2529292828 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 18 |
Hb_000174_140 |
0.2530022906 |
- |
- |
JHL23J11.8 [Jatropha curcas] |
| 19 |
Hb_007185_040 |
0.2534268594 |
- |
- |
PREDICTED: pyrroline-5-carboxylate reductase [Jatropha curcas] |
| 20 |
Hb_004976_050 |
0.2574198311 |
- |
- |
- |