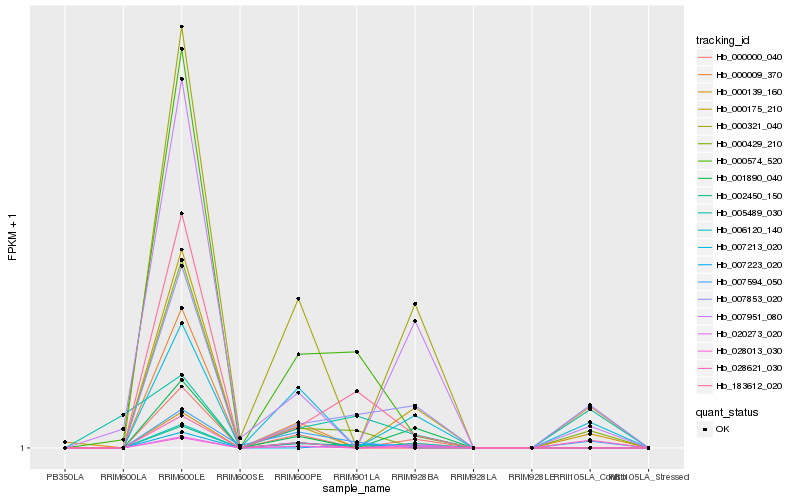
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007853_020 |
0.0 |
- |
- |
protein BOLA2 [Jatropha curcas] |
| 2 |
Hb_002450_150 |
0.2709312561 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000429_210 |
0.2768287381 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0089590 [Ricinus communis] |
| 4 |
Hb_007213_020 |
0.2931989628 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 5 |
Hb_183612_020 |
0.2968543971 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like [Nelumbo nucifera] |
| 6 |
Hb_007951_080 |
0.2974831158 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 7 |
Hb_007223_020 |
0.3135991631 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-3 [Jatropha curcas] |
| 8 |
Hb_000139_160 |
0.3227282306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000574_520 |
0.3255726368 |
- |
- |
PREDICTED: uncharacterized protein LOC105634842 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000175_210 |
0.3317123463 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 11 |
Hb_007594_050 |
0.3329048496 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000009_370 |
0.3347137365 |
- |
- |
hypothetical protein JCGZ_12284 [Jatropha curcas] |
| 13 |
Hb_028621_030 |
0.3347602712 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |
| 14 |
Hb_020273_020 |
0.3354682799 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_001890_040 |
0.3382964574 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 16 |
Hb_028013_030 |
0.3422574242 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000000_040 |
0.3455500505 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075-like [Jatropha curcas] |
| 18 |
Hb_006120_140 |
0.3457445915 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 19 |
Hb_000321_040 |
0.3473151965 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 5 [Jatropha curcas] |
| 20 |
Hb_005489_030 |
0.3482333959 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |