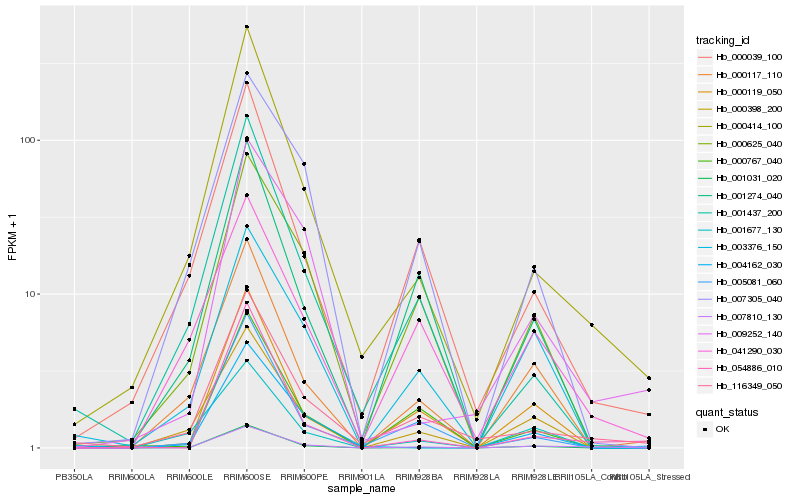
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007810_130 |
0.0 |
- |
- |
PREDICTED: ankyrin-1-like [Prunus mume] |
| 2 |
Hb_000414_100 |
0.1494998393 |
- |
- |
PREDICTED: uncharacterized protein LOC105643784 [Jatropha curcas] |
| 3 |
Hb_001031_020 |
0.1501745683 |
- |
- |
hypothetical protein VITISV_000631 [Vitis vinifera] |
| 4 |
Hb_116349_050 |
0.161816406 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_000398_200 |
0.1657765673 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 6 |
Hb_000119_050 |
0.1686149067 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 7 |
Hb_000117_110 |
0.1768098948 |
- |
- |
transferase, putative [Ricinus communis] |
| 8 |
Hb_054886_010 |
0.1776591326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000625_040 |
0.1782319472 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 10 |
Hb_005081_060 |
0.1842442871 |
- |
- |
hypothetical protein EUGRSUZ_C00833 [Eucalyptus grandis] |
| 11 |
Hb_000039_100 |
0.1842954707 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 12 |
Hb_001274_040 |
0.1865938854 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 13 |
Hb_000767_040 |
0.1879149168 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_009252_140 |
0.1899530563 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 15 |
Hb_007305_040 |
0.1908664512 |
- |
- |
Nodulin-26, putative [Ricinus communis] |
| 16 |
Hb_003376_150 |
0.1925768152 |
- |
- |
PREDICTED: peroxidase 21 [Jatropha curcas] |
| 17 |
Hb_041290_030 |
0.1945501719 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 18 |
Hb_001677_130 |
0.1957719978 |
- |
- |
hypothetical protein POPTR_0081s002101g, partial [Populus trichocarpa] |
| 19 |
Hb_001437_200 |
0.1957845342 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 20 |
Hb_004162_030 |
0.2005962364 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase-like [Jatropha curcas] |