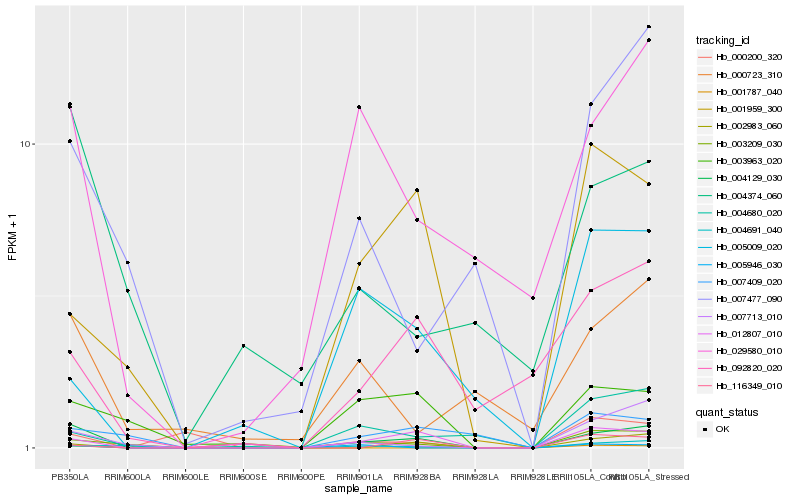
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004129_030 |
0.0 |
- |
- |
PREDICTED: cytochrome b561 domain-containing protein At2g30890-like isoform X2 [Populus euphratica] |
| 2 |
Hb_012807_010 |
0.2851677054 |
- |
- |
- |
| 3 |
Hb_029580_010 |
0.2955338584 |
- |
- |
PREDICTED: uncharacterized protein LOC105633393 [Jatropha curcas] |
| 4 |
Hb_002983_060 |
0.297837808 |
desease resistance |
Gene Name: NB-ARC |
Cc-nbs-lrr resistance protein, putative [Theobroma cacao] |
| 5 |
Hb_007713_010 |
0.3035245866 |
- |
- |
hypothetical protein JCGZ_18011 [Jatropha curcas] |
| 6 |
Hb_003963_020 |
0.3139204914 |
- |
- |
PREDICTED: uncharacterized protein LOC100256672 [Vitis vinifera] |
| 7 |
Hb_092820_020 |
0.3244049656 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_004374_060 |
0.3356363059 |
- |
- |
hypothetical protein POPTR_0003s13140g [Populus trichocarpa] |
| 9 |
Hb_003209_030 |
0.3372159404 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 10 |
Hb_001959_300 |
0.3378196577 |
- |
- |
PREDICTED: 3-isopropylmalate dehydratase large subunit [Jatropha curcas] |
| 11 |
Hb_007477_090 |
0.3396934771 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 12 |
Hb_000723_310 |
0.3406888701 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_004680_020 |
0.3448209138 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105789475 [Gossypium raimondii] |
| 14 |
Hb_004691_040 |
0.3464639124 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 15 |
Hb_001787_040 |
0.3488266259 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_005946_030 |
0.3490683284 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 17 |
Hb_007409_020 |
0.3561974214 |
- |
- |
- |
| 18 |
Hb_005009_020 |
0.3592004711 |
- |
- |
- |
| 19 |
Hb_000200_320 |
0.3622528609 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein JCGZ_09311 [Jatropha curcas] |
| 20 |
Hb_116349_010 |
0.3627723651 |
- |
- |
PREDICTED: uncharacterized protein LOC105763153 [Gossypium raimondii] |