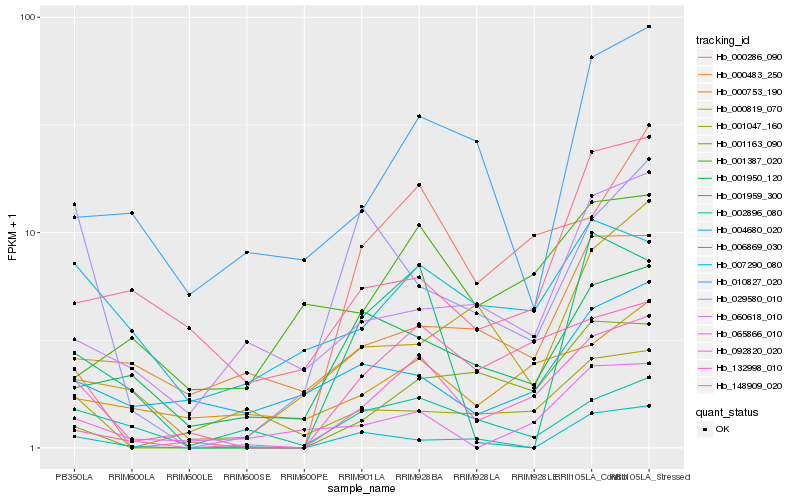
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_092820_020 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_000286_090 |
0.233997735 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_00197 [Jatropha curcas] |
| 3 |
Hb_001163_090 |
0.2380688709 |
- |
- |
hypothetical protein POPTR_0009s14920g [Populus trichocarpa] |
| 4 |
Hb_132998_010 |
0.2439280352 |
- |
- |
hypothetical protein JCGZ_00197 [Jatropha curcas] |
| 5 |
Hb_000753_190 |
0.2568675536 |
- |
- |
PREDICTED: D-aminoacyl-tRNA deacylase [Populus euphratica] |
| 6 |
Hb_065866_010 |
0.2577009981 |
- |
- |
PREDICTED: hydroxyacyl-thioester dehydratase type 2, mitochondrial [Fragaria vesca subsp. vesca] |
| 7 |
Hb_002896_080 |
0.2619258332 |
- |
- |
- |
| 8 |
Hb_029580_010 |
0.2624756799 |
- |
- |
PREDICTED: uncharacterized protein LOC105633393 [Jatropha curcas] |
| 9 |
Hb_006869_030 |
0.2641184694 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_060618_010 |
0.2670365714 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 11 |
Hb_148909_020 |
0.2717306952 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 12 |
Hb_001950_120 |
0.2729832158 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004680_020 |
0.2787921216 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105789475 [Gossypium raimondii] |
| 14 |
Hb_001047_160 |
0.2790238265 |
- |
- |
- |
| 15 |
Hb_000483_250 |
0.279237231 |
- |
- |
PREDICTED: uncharacterized protein LOC105803540 [Gossypium raimondii] |
| 16 |
Hb_010827_020 |
0.2825367317 |
- |
- |
60S ribosomal protein L17B [Hevea brasiliensis] |
| 17 |
Hb_000819_070 |
0.2827707791 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |
| 18 |
Hb_001959_300 |
0.284245373 |
- |
- |
PREDICTED: 3-isopropylmalate dehydratase large subunit [Jatropha curcas] |
| 19 |
Hb_001387_020 |
0.2865979693 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 20 |
Hb_007290_080 |
0.2892985834 |
- |
- |
hypothetical protein CISIN_1g047098mg [Citrus sinensis] |