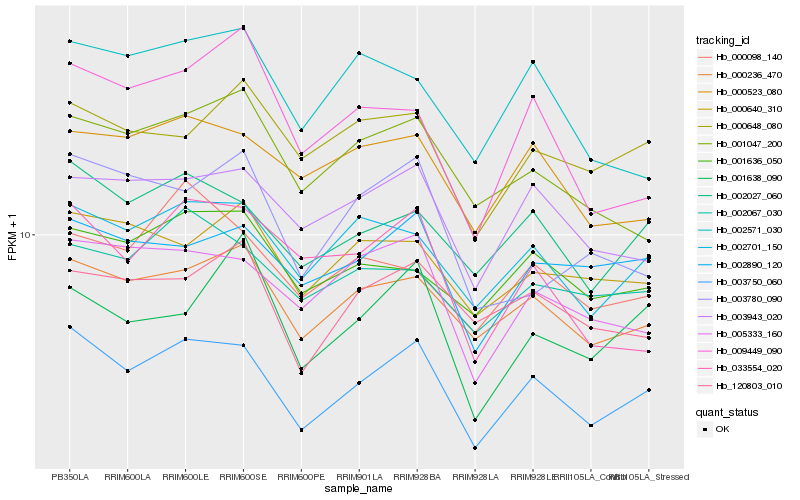
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003750_060 |
0.0 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002027_060 |
0.1066264201 |
- |
- |
PREDICTED: exosome complex component RRP4 [Jatropha curcas] |
| 3 |
Hb_000236_470 |
0.1124039621 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 4 |
Hb_002701_150 |
0.1142926497 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005333_160 |
0.1144124369 |
- |
- |
hypothetical protein B456_009G230200 [Gossypium raimondii] |
| 6 |
Hb_003943_020 |
0.1238529634 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002890_120 |
0.1239191944 |
- |
- |
Nucleic acid binding,RNA binding isoform 1 [Theobroma cacao] |
| 8 |
Hb_033554_020 |
0.127609023 |
transcription factor |
TF Family: BES1 |
PREDICTED: beta-amylase 8 [Jatropha curcas] |
| 9 |
Hb_000523_080 |
0.1306744243 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 10 |
Hb_009449_090 |
0.1310618452 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001047_200 |
0.1323127634 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 12 |
Hb_001636_050 |
0.1345675105 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 13 |
Hb_001638_090 |
0.1348598561 |
- |
- |
GRF1-INTERACTING FACTOR 2 family protein [Populus trichocarpa] |
| 14 |
Hb_003780_090 |
0.135034857 |
- |
- |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 15 |
Hb_120803_010 |
0.1351215902 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein [Malus domestica] |
| 16 |
Hb_000640_310 |
0.1366774571 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 17 |
Hb_002067_030 |
0.1369341665 |
- |
- |
PREDICTED: poly(ADP-ribose) glycohydrolase 1-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_002571_030 |
0.1379078746 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 19 |
Hb_000098_140 |
0.1384591802 |
- |
- |
hypothetical protein JCGZ_04989 [Jatropha curcas] |
| 20 |
Hb_000648_080 |
0.138476492 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |