| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
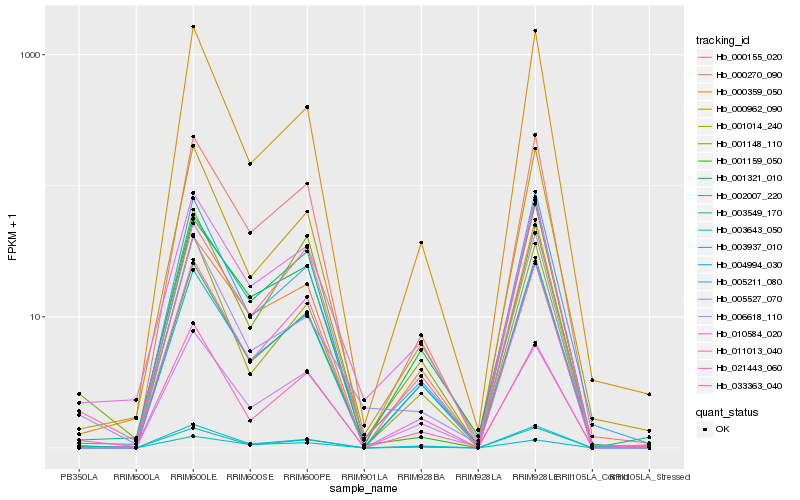
Hb_003549_170 |
0.0 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa b, chloroplastic [Jatropha curcas] |
| 2 |
Hb_010584_020 |
0.1205568725 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 3 |
Hb_021443_060 |
0.1232578469 |
- |
- |
PTH-1 family protein [Populus trichocarpa] |
| 4 |
Hb_003937_010 |
0.1238037454 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 5 |
Hb_033363_040 |
0.1269548691 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 6 |
Hb_000962_090 |
0.1295714804 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 7 |
Hb_005527_070 |
0.1310921983 |
- |
- |
short-chain dehydrogenase/reductase family protein [Populus trichocarpa] |
| 8 |
Hb_004994_030 |
0.1324718387 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 9 |
Hb_000155_020 |
0.1336444565 |
- |
- |
PREDICTED: triose phosphate/phosphate translocator TPT, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_002007_220 |
0.1362124175 |
- |
- |
PREDICTED: signal recognition particle 43 kDa protein, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005211_080 |
0.1362456963 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |
| 12 |
Hb_003643_050 |
0.1372133909 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Fragaria vesca subsp. vesca] |
| 13 |
Hb_001159_050 |
0.1384950007 |
- |
- |
PREDICTED: uncharacterized protein LOC105645952 [Jatropha curcas] |
| 14 |
Hb_000270_090 |
0.1386158176 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 15 |
Hb_001014_240 |
0.139354144 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.1 [Jatropha curcas] |
| 16 |
Hb_006618_110 |
0.1396705675 |
- |
- |
- |
| 17 |
Hb_001321_010 |
0.1402016982 |
- |
- |
HIGH-CHLOROPHYLL-FLUORESCENCE 101 family protein [Populus trichocarpa] |
| 18 |
Hb_001148_110 |
0.1405211798 |
- |
- |
- |
| 19 |
Hb_011013_040 |
0.1411350621 |
transcription factor |
TF Family: C2C2-YABBY |
unknown [Populus trichocarpa] |
| 20 |
Hb_000359_050 |
0.141982606 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |