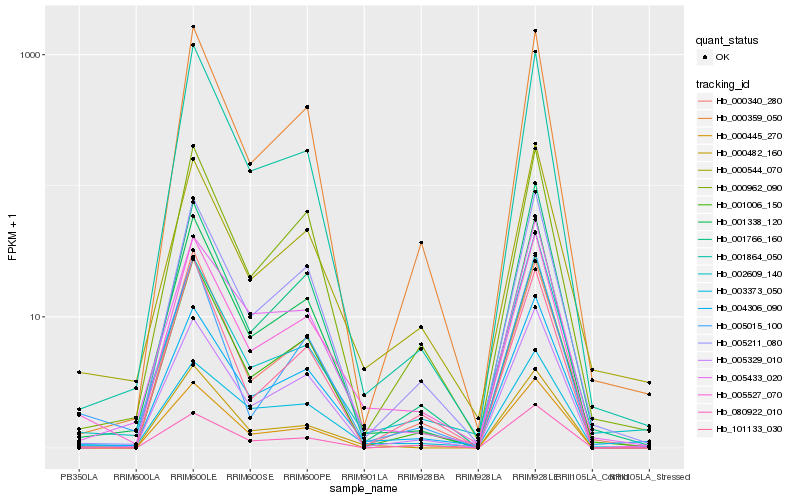
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005527_070 |
0.0 |
- |
- |
short-chain dehydrogenase/reductase family protein [Populus trichocarpa] |
| 2 |
Hb_001338_120 |
0.0803839213 |
- |
- |
hypothetical chloroplast RF68 (chloroplast) [Ipomoea batatas] |
| 3 |
Hb_004306_090 |
0.0816937972 |
- |
- |
PREDICTED: uncharacterized protein LOC105644392 isoform X2 [Jatropha curcas] |
| 4 |
Hb_005211_080 |
0.0933900732 |
- |
- |
hypothetical protein POPTR_0010s13740g [Populus trichocarpa] |
| 5 |
Hb_000544_070 |
0.0955756952 |
- |
- |
Peptidyl-prolyl cis-trans isomerase FKBP17-2 [Morus notabilis] |
| 6 |
Hb_005329_010 |
0.0959144611 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 7 |
Hb_001006_150 |
0.097923659 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_000359_050 |
0.1015295076 |
- |
- |
Oxygen-evolving enhancer protein 2, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000962_090 |
0.1017250025 |
- |
- |
omega-6 fatty acid desaturase [Hevea brasiliensis] |
| 10 |
Hb_000445_270 |
0.1031974377 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_003373_050 |
0.1082229018 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 12 |
Hb_001864_050 |
0.1084698722 |
- |
- |
chloroplast oxygen-evolving enhancer protein [Manihot esculenta] |
| 13 |
Hb_000482_160 |
0.1106565764 |
- |
- |
hypothetical protein JCGZ_07670 [Jatropha curcas] |
| 14 |
Hb_005433_020 |
0.1138077969 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 15 |
Hb_005015_100 |
0.1158787312 |
- |
- |
PREDICTED: uncharacterized protein LOC105636541 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001766_160 |
0.1161842959 |
- |
- |
Serine/threonine-protein kinase SAPK1 [Gossypium arboreum] |
| 17 |
Hb_000340_280 |
0.1170034545 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_080922_010 |
0.1181302888 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 19 |
Hb_101133_030 |
0.1190956153 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_002609_140 |
0.1191799894 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic isoform X1 [Jatropha curcas] |